8HT7
 
 | | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), GLN-ALA-GLN-ALA-THR-ILE-SER-PHE-PRO-LYS-ARG-LYS-LEU-SER-TRP | | Authors: | Liu, C, Zhu, G, Geng, Y, Xu, N. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex.
Int.J.Biol.Macromol., 260, 2024
|
|
6MQC
 
 | |
6MPH
 
 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MQM
 
 | |
6N16
 
 | |
6N1W
 
 | |
6NF2
 
 | |
6MQE
 
 | |
6MQS
 
 | |
6MQR
 
 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6N1V
 
 | |
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
9IVB
 
 | |
3VOD
 
 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
9JVZ
 
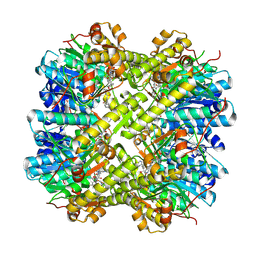 | | CryoEM structure of M. tuberculosis ClpP1P2 bound to bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, W, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
6UIN
 
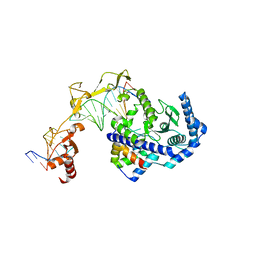 | | Role of Beta-hairpin motifs in the DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*NP*NP*NP*NP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | Tethering-facilitated DNA 'opening' and complementary roles of beta-hairpin motifs in the Rad4/XPC DNA damage sensor protein
Nucleic Acids Res., 48, 2021
|
|
6RAB
 
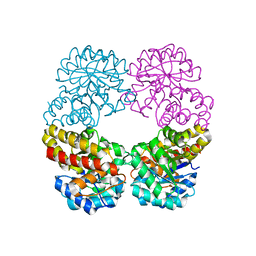 | |
6RD1
 
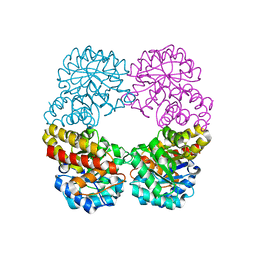 | | Ruminococcus gnavus sialic acid aldolase catalytic lysine mutant in complex with sialic acid | | Descriptor: | 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, Putative N-acetylneuraminate lyase | | Authors: | Owen, C.D, Bell, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-04-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut.
Nat Microbiol, 4, 2019
|
|
6IZK
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
9DI9
 
 | | Rat branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (3M)-6-fluoro-3-(2,4,5-trifluoro-3-methoxyphenyl)-1-benzothiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, Branched-chain alpha-ketoacid dehydrogenase kinase, ... | | Authors: | Liu, S. | | Deposit date: | 2024-09-05 | | Release date: | 2024-11-27 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Discovery of First Branched-Chain Ketoacid Dehydrogenase Kinase (BDK) Inhibitor Clinical Candidate PF-07328948.
J.Med.Chem., 68, 2025
|
|
6RB7
 
 | | Ruminococcus gnavus sialic acid aldolase catalytic lysine mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Bell, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-04-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut.
Nat Microbiol, 4, 2019
|
|
6QSU
 
 | | Helicobacter pylori urease with BME bound in the active site | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2019-02-22 | | Release date: | 2021-01-20 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
1KBR
 
 | |
5JXU
 
 | | Structural basis for the catalytic activity of Thermomonospora curvata heme-containing DyP-type peroxidase. | | Descriptor: | Dyp-type peroxidase family, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ramyar, K.X, Carlson, E.A, Li, P, Geisbrecht, B.V. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Identification of Surface-Exposed Protein Radicals and A Substrate Oxidation Site in A-Class Dye-Decolorizing Peroxidase from Thermomonospora curvata.
ACS Catal, 6, 2016
|
|
3TPX
 
 | |
