6BAN
 
 | |
5JJA
 
 | | Crystal structure of a PP2A B56gamma/BubR1 complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Protein Cell, 7, 2016
|
|
6VBN
 
 | | Crystal Structure of hTDO2 bound to inhibitor GNE1 | | Descriptor: | 1,5-anhydro-2,3-dideoxy-3-[(5S)-5H-imidazo[5,1-a]isoindol-5-yl]-D-threo-pentitol, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Implementation of the CYP Index for the Design of Selective Tryptophan-2,3-dioxygenase Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
5K24
 
 | |
5JYK
 
 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
6VQL
 
 | |
5K25
 
 | |
5K23
 
 | |
6WH9
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
3FW5
 
 | |
3FW4
 
 | |
6W7S
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 2G10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G10 (Fab heavy chain), 2G10 (Fab light chain), ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
8K4Q
 
 | |
2O3C
 
 | | Crystal structure of zebrafish Ape | | Descriptor: | APEX nuclease 1, LEAD (II) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
6IHB
 
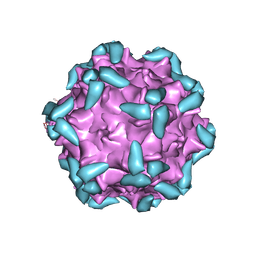 | | Adeno-Associated Virus 2 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
2O3H
 
 | | Crystal structure of the human C65A Ape | | Descriptor: | ACETATE ION, DNA-(apurinic or apyrimidinic site) lyase, SAMARIUM (III) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
6IH9
 
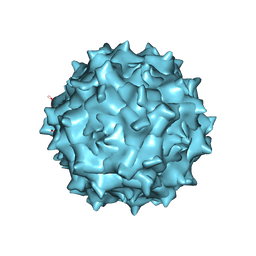 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
6KI8
 
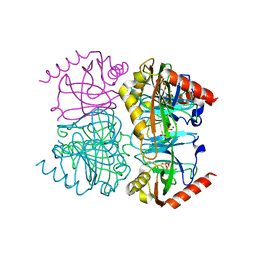 | | Pyrophosphatase mutant K149R from Acinetobacter baumannii | | Descriptor: | DIPHOSPHATE, Inorganic pyrophosphatase, MAGNESIUM ION | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
6WAA
 
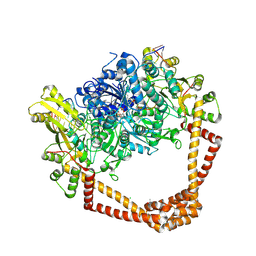 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and compound 34 (7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Topoisomerase Inhibitors Addressing Fluoroquinolone Resistance in Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6KJW
 
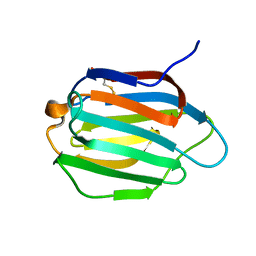 | | Galectin-13 variant C136S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
2I03
 
 | | Crystal structure of human dipeptidyl peptidase 4 (DPP IV) with potent alkynyl cyanopyrrolidine (ABT-279) | | Descriptor: | 2-[4-({2-[(2S,5R)-2-(AMINOMETHYL)-5-ETHYNYLPYRROLIDIN-1-YL]-2-OXOETHYL}AMINO)-4-METHYLPIPERIDIN-1-YL]ISONICOTINIC ACID, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Madar, D.J. | | Deposit date: | 2006-08-09 | | Release date: | 2006-12-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]- 4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes.
J.Med.Chem., 49, 2006
|
|
6WHO
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHQ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WSJ
 
 | |
