4FL2
 
 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
4FO1
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lincosamide resistance protein | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo
TO BE PUBLISHED
|
|
4FO9
 
 | | Crystal structure of the E3 SUMO Ligase PIAS2 | | Descriptor: | E3 SUMO-protein ligase PIAS2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Hu, J, Dobrovetsky, E, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the E3 SUMO Ligase PIAS2
to be published
|
|
4F9K
 
 | | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A | | Descriptor: | cAMP-dependent protein kinase type I-beta regulatory subunit | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Shastry, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-18 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A
To be Published
|
|
4FDF
 
 | |
4FCZ
 
 | | Crystal Structure of Toluene-tolerance protein from Pseudomonas putida (strain KT2440), Northeast Structural Genomics Consortium (NESG) Target PpR99 | | Descriptor: | Toluene-tolerance protein | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-25 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Northeast Structural Genomics Consortium Target PpR99
To be Published
|
|
4FEZ
 
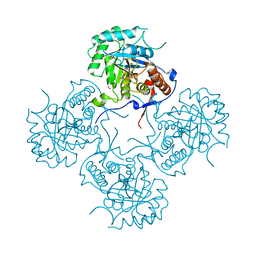 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant.
To be Published
|
|
4FFS
 
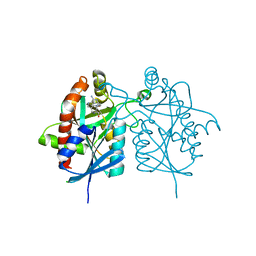 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Helicobacter pylori with butyl-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Haapalainen, A.M, Rinaldo-Matthis, A, Brown, R.L, Norris, G.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Picomolar Transition State Analogue Inhibitor of MTAN as a Specific Antibiotic for Helicobacter pylori.
Biochemistry, 51, 2012
|
|
4FFV
 
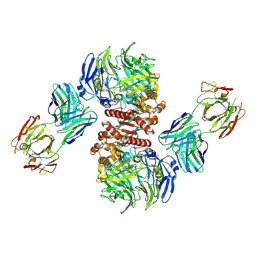 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab | | Descriptor: | 11A19 Fab heavy chain, 11A19 Fab light chain, Dipeptidyl peptidase 4 | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
4FH5
 
 | |
4FMW
 
 | | Crystal structure of methyltransferase domain of human RNA (guanine-9-) methyltransferase domain containing protein 2 | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, RNA (guanine-9-)-methyltransferase domain-containing protein 2, ... | | Authors: | Dong, A, Zeng, H, Loppnau, P, Tempel, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of methyltransferase domain of human RNA (guanine-9-) methyltransferase domain containing protein 2
TO BE PUBLISHED
|
|
2Y9W
 
 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
2YMA
 
 | | X-ray structure of the Yos9 dimerization domain | | Descriptor: | PROTEIN OS-9 HOMOLOG | | Authors: | Hanna, J, Schuetz, A, Zimmermann, F, Behlke, J, Sommer, T, Heinemann, U. | | Deposit date: | 2011-06-07 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structural and Biochemical Basis of Yos9 Protein Dimerization and Possible Contribution to Self-Association of 3-Hydroxy-3-Methylglutaryl-Coenzyme a Reductase Degradation Ubiquitin-Ligase Complex.
J.Biol.Chem., 287, 2012
|
|
4UD8
 
 | | AtBBE15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Daniel, B, Steiner, B, Pavkov-Keller, T, Dordic, A, Gutmann, A, Sensen, C.W, Nidetzky, B, van der Graaff, E, Wallner, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Oxidation of Monolignols by Members of the Berberine Bridge Enzyme Family Suggests a Role in Cell Wall Metabolism.
J.Biol.Chem., 290, 2015
|
|
4V7G
 
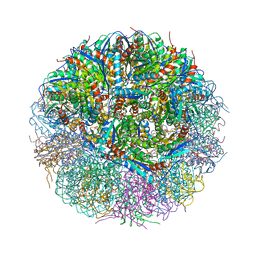 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | Deposit date: | 2009-09-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4V5M
 
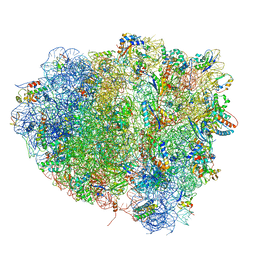 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
4V5A
 
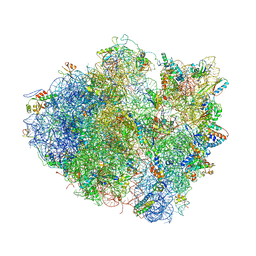 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
4UYK
 
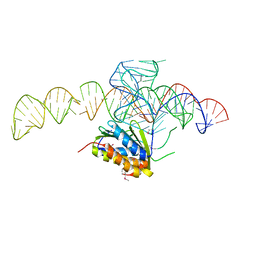 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
4V0Z
 
 | |
4V4O
 
 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
4V9F
 
 | |
4V9O
 
 | |
4UWU
 
 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 7) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
4URS
 
 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
4UQV
 
 | |
