1LJZ
 
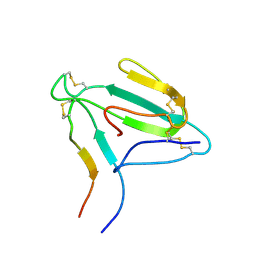 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Eisenstein, M, Chill, J.H, Anglister, J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1UCB
 
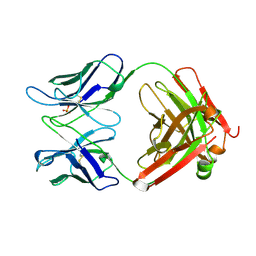 | |
1KZ7
 
 | | Crystal Structure of the DH/PH Fragment of Murine Dbs in Complex with the Placental Isoform of Human Cdc42 | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
6EZN
 
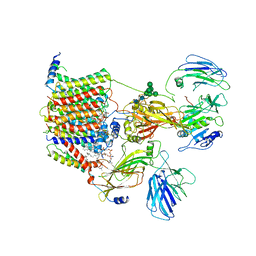 | | Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Kowal, J, Eyring, J, Ngwa, E.M, Aebi, M, Locher, K.P. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation.
Science, 359, 2018
|
|
5XRM
 
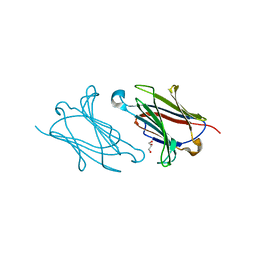 | |
6TK1
 
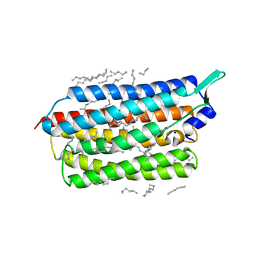 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 20ms structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
2IFT
 
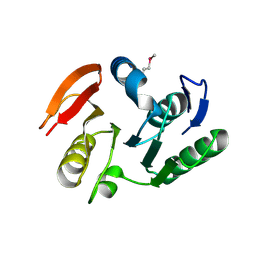 | | Crystal structure of putative methylase HI0767 from Haemophilus influenzae. NESG target IR102. | | Descriptor: | Putative methylase HI0767 | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Shastry, R, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the putative methylase HI0767 from Haemophilus influenzae.
To be Published
|
|
2IGQ
 
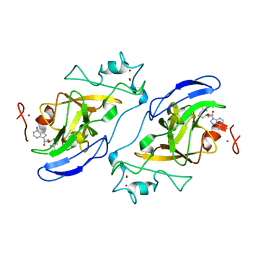 | | Human euchromatic histone methyltransferase 1 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, euchromatic histone methyltransferase 1 | | Authors: | Min, J, Wu, H, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-23 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
5XPV
 
 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
1U5B
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, GLYCEROL, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-07-27 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
4CPX
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl ~{N}-[(2~{S})-3,3-dimethyl-1-[2-[3-[(3~{R},6~{S},10~{Z})-3-oxidanyl-4,7-bis(oxidanylidene)-6-propan-2-yl-5,8-diazabicyclo[11.2.2]heptadeca-1(16),10,13(17),14-tetraen-3-yl]propyl]-2-[(4-thiophen-2-ylphenyl)methyl]hydrazinyl]-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of P1'-functionalized macrocyclic transition-state mimicking HIV-1 protease inhibitors encompassing a tertiary alcohol.
J. Med. Chem., 57, 2014
|
|
6TPM
 
 | | Crystal structure of AmpC from E.coli with Relebactam (MK-7655) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Investigations of the Inhibition of Escherichia coli AmpC beta-Lactamase by Diazabicyclooctanes.
Antimicrob.Agents Chemother., 65, 2021
|
|
5XSQ
 
 | | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35 | | Authors: | Zhu, T, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-06-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide Reveals a Conserved Drug Target for Filovirus
J. Virol., 91, 2017
|
|
6TRD
 
 | | Cryo- EM structure of the Thermosynechococcus elongatus photosystem I in the presence of cytochrome c6 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Koelsch, A, Radon, C, Baumert, A, Buerger, J, Mielke, T, Lisdat, F, Zouni, A, Wendler, P. | | Deposit date: | 2019-12-18 | | Release date: | 2020-09-16 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I
Curr.Opin.Struct.Biol., 2, 2020
|
|
5XRH
 
 | | Galectin-10/Charcot-Leyden crystal protein crystal structure | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
6TK5
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 800fs+2ps structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
5XRN
 
 | |
6TK7
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: Dark structure in acidic conditions | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
8S5R
 
 | | Structure of the Chlamydia pneumoniae effector SemD | | Descriptor: | Effector SemD | | Authors: | Kocher, F, Applegate, V, Reiners, J, Port, A, Spona, D, Haensch, S, Smits, S.H, Hegemann, J, Moelleken, K. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Chlamydia pneumoniae effector SemD exploits its host's endocytic machinery by structural and functional mimicry.
Nat Commun, 15, 2024
|
|
8S5T
 
 | | Structure of SemD in complex | | Descriptor: | Effector SemD, Neural Wiskott-Aldrich syndrome protein | | Authors: | Kocher, F, Applegate, V, Port, A, Reiners, J, Spona, D, Haensch, S, Smits, S.H, Hegemann, J, Moelleken, K. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Chlamydia pneumoniae effector SemD exploits its host's endocytic machinery by structural and functional mimicry.
Nat Commun, 15, 2024
|
|
1KZG
 
 | | DbsCdc42(Y889F) | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
5XWV
 
 | |
6TTF
 
 | | PKM2 in complex with Compound 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-hydroxynaphthalene-1-sulfonamide, Pyruvate kinase PKM | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
4CYJ
 
 | | Chaetomium thermophilum Pan2:Pan3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
