4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6K9N
 
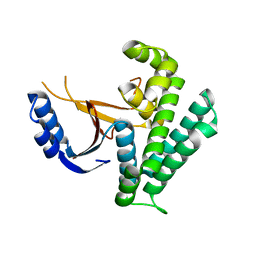 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
7XWX
 
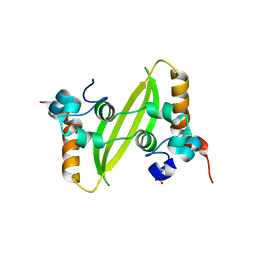 | | Crystal structure of SARS-CoV-2 N-CTD | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XX1
 
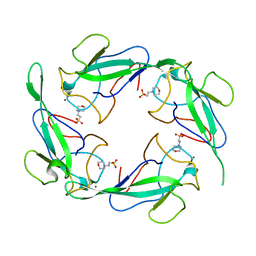 | | Crystal structure of SARS-CoV-2 N-NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XWZ
 
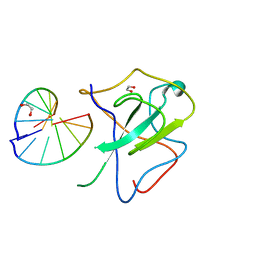 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6KBE
 
 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
7XUR
 
 | |
6L7C
 
 | | CsgFG complex with substrate CsgAN6 peptide in Curli biogenesis system | | Descriptor: | CsgF, Curli production assembly/transport protein CsgG, Major curlin subunit CsgA | | Authors: | Yan, Z.F, Yin, M, Chen, J.N, Li, X.M. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Assembly and substrate recognition of curli biogenesis system.
Nat Commun, 11, 2020
|
|
7XQP
 
 | | PSI-LHCI-LHCII-Lhcb9 supercomplex of Physcomitrella patens | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhang, S, Tang, K.L, Li, X.Y, Wang, W.D, Yan, Q.J, Shen, L.L, Kuang, T.Y, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
7YW2
 
 | | Crystal structure of tRNA 2'-phosphotransferase from Mus musculus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
7YW3
 
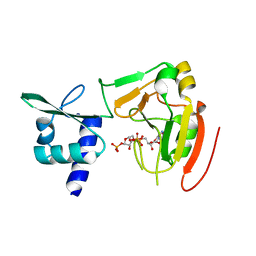 | | Crystal structure of tRNA 2'-phosphotransferase from Homo sapiens | | Descriptor: | 1,2-ETHANEDIOL, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate, tRNA 2'-phosphotransferase 1 | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
7YW4
 
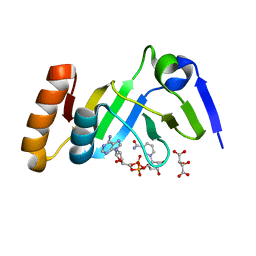 | |
7W0S
 
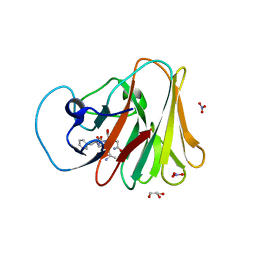 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0Q
 
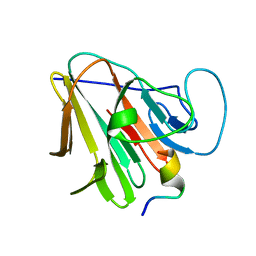 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0T
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
3GA3
 
 | | Crystal structure of the C-terminal domain of human MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, MDA5, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2009-02-16 | | Release date: | 2009-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the C-terminal domain of human MDA5
To be Published
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
8TRG
 
 | | Structure of full-length LexA bound to a RecA filament | | Descriptor: | DNA (27-MER), LexA repressor, MAGNESIUM ION, ... | | Authors: | Cory, M.B, Li, A, Kohli, R.M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The LexA-RecA* structure reveals a cryptic lock-and-key mechanism for SOS activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5ZXK
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6A7B
 
 | | AKR1C3 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
7AHU
 
 | | Anti-FX Fab of mim8 in complex with human FXa | | Descriptor: | Anti-FX Fab of mim8 heavy chain, Anti-FX Fab of mim8 light chain, CALCIUM ION, ... | | Authors: | Johansson, E. | | Deposit date: | 2020-09-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A factor VIIIa-mimetic bispecific antibody, Mim8, ameliorates bleeding upon severe vascular challenge in hemophilia A mice.
Blood, 138, 2021
|
|
7AHV
 
 | | Anti-FIXa Fab of mim8 in complex with human FIXa | | Descriptor: | CALCIUM ION, Coagulation factor IX, L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, ... | | Authors: | Johansson, E. | | Deposit date: | 2020-09-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | A factor VIIIa-mimetic bispecific antibody, Mim8, ameliorates bleeding upon severe vascular challenge in hemophilia A mice.
Blood, 138, 2021
|
|
