2KL2
 
 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KA5
 
 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
2VIS
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, (ESCAPE) MUTANT WITH THR 131 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
2VIT
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, MUTANT WITH THR 155 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, LAMBDA), ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
3IMT
 
 | | Transthyretin in complex with (E)-4-(4-aminostyryl)-2,6-dibromophenol | | Descriptor: | 4-[(E)-2-(4-aminophenyl)ethenyl]-2,6-dibromophenol, Transthyretin | | Authors: | Connelly, S, Wilson, I. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
7KSG
 
 | | SARS-CoV-2 spike in complex with nanobodies E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 glycoprotein, Spike glycoprotein | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-22 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7ZLK
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS114 and ACS122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS114 heavy chain, ... | | Authors: | van Schooten, J, Ozorowski, G, Ward, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7Z3A
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS101 heavy chain, ... | | Authors: | van Schooten, J, Ward, A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7B17
 
 | | SARS-CoV-spike RBD bound to two neutralising nanobodies. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama,SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B14
 
 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B18
 
 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
3F58
 
 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 12-RESIDUE CYCLIC PEPTIDE (INCLUDING RESIDUES 315-324 OF HIV-1 GP120 (MN ISOLATE); H315S MUTATION | | Descriptor: | PROTEIN (CYCLIC PEPTIDE (GP120)), PROTEIN (IMMUNOGLOBULIN GAMMA I (58.2)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-23 | | Release date: | 1999-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
6GK8
 
 | | Crystal structure of anti-tau antibody dmCBTAU-28.1, double mutant (S32R, E35K) of CBTAU-28.1, in complex with Tau peptide A7731 (residues 52-71) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-28.1(S32R;E35K), TAU PEPTIDE A7731 (RESIDUES 52-71) | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
6GK7
 
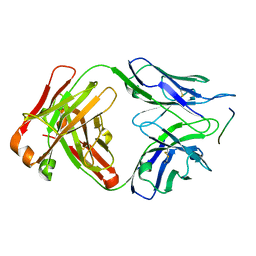 | | Crystal structure of anti-tau antibody dmCBTAU-27.1, double mutant (S31Y, T100I) of CBTAU-27.1, in complex with Tau peptide A8119B (residues 299-318) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-27.1(S31Y,T100I), HUMAN TAU PEPTIDE A8119 RESIDUES 299-318 | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
9MII
 
 | |
9MI0
 
 | |
9MIA
 
 | |
9MIH
 
 | |
9MIB
 
 | |
2OOK
 
 | |
2OOC
 
 | |
2QAD
 
 | | Structure of tyrosine-sulfated 412d antibody complexed with HIV-1 YU2 gp120 and CD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Huang, C.-C, Tang, M, Robinson, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
2QMQ
 
 | |
2Q8U
 
 | |
