7TB6
 
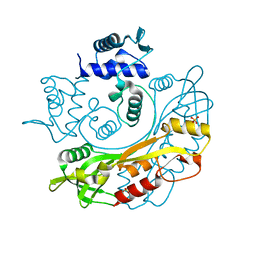 | | Structure of S. maltophilia CapW | | Descriptor: | S. maltophilia CapW, SULFATE ION | | Authors: | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
7TB5
 
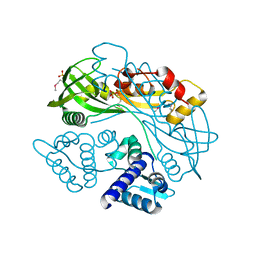 | | Structure of P. aeruginosa PA17 CapW | | Descriptor: | SULFATE ION, WYL domain-containing protein | | Authors: | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
5MSI
 
 | |
3MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Descriptor: | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Authors: | Deluca, C.I, Davies, P.L, Ye, Q, Jia, Z. | | Deposit date: | 1997-09-17 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice.
J.Mol.Biol., 275, 1998
|
|
7MSI
 
 | |
6MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Descriptor: | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Authors: | Deluca, C.I, Davies, P.L, Ye, Q, Jia, Z. | | Deposit date: | 1997-09-17 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice.
J.Mol.Biol., 275, 1998
|
|
3HAG
 
 | | Crystal structure of the Hepatitis E Virus-like Particle | | Descriptor: | Capsid protein | | Authors: | Guu, T.S.Y, Liu, Z, Ye, Q, Mata, D.A, Li, K, Yin, C, Zhang, J, Tao, Y.J. | | Deposit date: | 2009-05-01 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2MSI
 
 | |
4MSI
 
 | |
2MOK
 
 | | holo_FldA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2014-04-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR study of YqcA from Escherichia coli
To be Published
|
|
4E03
 
 | | Structure of ParF-ADP form 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4DZZ
 
 | | Structure of ParF-ADP, crystal form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E09
 
 | | Structure of ParF-AMPPCP, I422 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF, SULFATE ION | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E07
 
 | | ParF-AMPPCP-C2221 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
6DXK
 
 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
7TQD
 
 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TO3
 
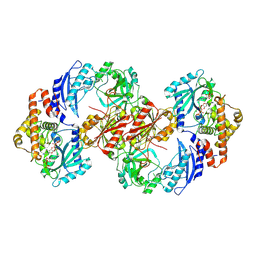 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
2P98
 
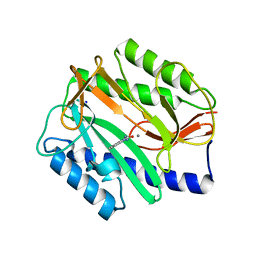 | | E. coli methionine aminopeptidase monometalated with inhibitor YE7 | | Descriptor: | IMIDAZO[2,1-A]ISOQUINOLINE-2-CARBOHYDRAZIDE, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
2P9A
 
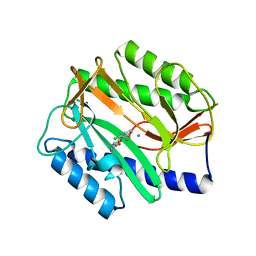 | | E. coli methionine aminopeptidase dimetalated with inhibitor YE6 | | Descriptor: | 5-(2-chlorophenyl)furan-2-carbohydrazide, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
2P99
 
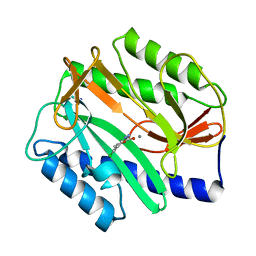 | | E. coli methionine aminopeptidase monometalated with inhibitor YE6 | | Descriptor: | 5-(2-chlorophenyl)furan-2-carbohydrazide, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
5DYJ
 
 | | Mysosin heavy chain kinase A catalytic domain mutant - D663A | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Myosin heavy chain kinase A, ... | | Authors: | van Staalduinen, L.M, Yang, Y, Jia, Z. | | Deposit date: | 2015-09-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Dictyostelium Myosin-II Heavy Chain Kinase A (MHCK-A) alpha-kinase domain apoenzyme reveals a novel autoinhibited conformation.
Sci Rep, 6, 2016
|
|
5JDK
 
 | |
5WSN
 
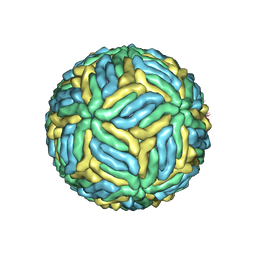 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
6FS2
 
 | | MCL1 in complex with indole acid ligand | | Descriptor: | 7-(2-methylphenyl)-3-[3-(5,6,7,8-tetrahydronaphthalen-1-yloxy)propyl]-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6FS1
 
 | | MCL1 in complex with an indole acid ligand | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-[(1,5-dimethylpyrazol-3-yl)methylsulfanylmethyl]-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Kasmirski, S, Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
