5TFA
 
 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dUTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFG
 
 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with 5-methyl-UTP | | Descriptor: | 5-Methyluridine triphosphate, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFB
 
 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with 7-methyl-GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFI
 
 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFC
 
 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with GTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFD
 
 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
6KOL
 
 | | Crystal structure of auracyanin from photosynthetic bacterium Roseiflexus castenholzii | | Descriptor: | Blue (Type 1) copper domain protein, CHLORIDE ION, COPPER (II) ION | | Authors: | Wang, C, Zhang, C.Y, Min, Z.Z, Xin, Y.Y, Xu, X.L. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Structural basis underlying the electron transfer features of a blue copper protein auracyanin from the photosynthetic bacterium Roseiflexus castenholzii.
Photosyn. Res., 143, 2020
|
|
6L9S
 
 | | Crystal structure of Na-dithionite reduced auracyanin from photosynthetic bacterium Roseiflexus castenholzii | | Descriptor: | Blue (Type 1) copper domain protein, COPPER (I) ION | | Authors: | Wang, C, Zhang, C.Y, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis underlying the electron transfer features of a blue copper protein auracyanin from the photosynthetic bacterium Roseiflexus castenholzii.
Photosyn. Res., 143, 2020
|
|
5Y5W
 
 | |
7DL9
 
 | | Crystal structure of nucleoside transporter NupG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Nucleoside permease NupG | | Authors: | Wang, C, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for substrate recognition by the bacterial nucleoside transporter NupG.
J.Biol.Chem., 296, 2021
|
|
7DLA
 
 | |
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
7UVP
 
 | |
6UK1
 
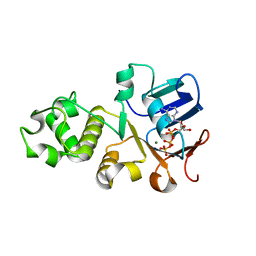 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
5YVN
 
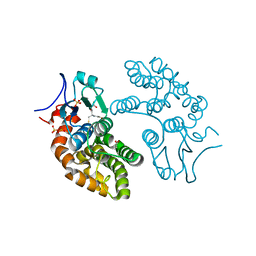 | | Human Glutathione Transferase Omega1 | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase omega-1, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
5YVO
 
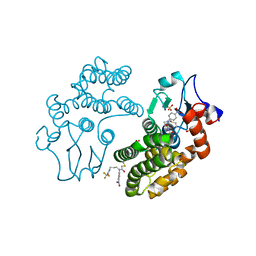 | | Human Glutathione Transferase Omega1 covalently bound to ML175 inhibitor | | Descriptor: | ACETATE ION, Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
6ZXV
 
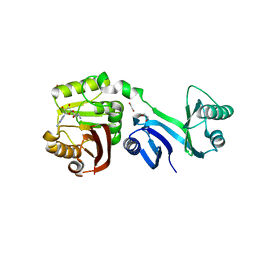 | |
6ZXY
 
 | |
6ZXW
 
 | |
6RJK
 
 | | Structure of virulence factor SghA from Agrobacterium tumefaciens | | Descriptor: | Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RK2
 
 | | Complex structure of virulence factor SghA mutant with its substrate SAG | | Descriptor: | 2-(alpha-L-altropyranosyloxy)benzoic acid, Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJM
 
 | | Complex structure of virulence factor SghA and its hydrolysis product glucose | | Descriptor: | Beta-glucosidase, alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
