4PID
 
 | | Crystal structure of human adenovirus 2 protease with a weak pyrimidine nitrile inhibitor | | Descriptor: | ACETATE ION, N-benzyl-2-[(Z)-iminomethyl]pyrimidine-5-carboxamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIQ
 
 | | Crystal structure of human adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | N-[(3,5-dichlorophenyl)acetyl]-L-threonyl-N-[(2Z)-2-iminoethyl]glycinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIS
 
 | | Crystal structure of human adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | N~2~-[(2R)-2-(3,5-dichlorophenyl)-2-(dimethylamino)acetyl]-N-({2-[(Z)-iminomethyl]pyrimidin-4-yl}methyl)-L-isoleucinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
8R12
 
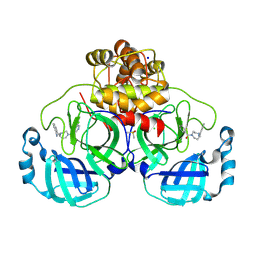 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | Descriptor: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R11
 
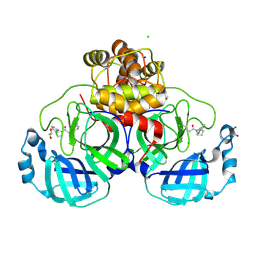 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R14
 
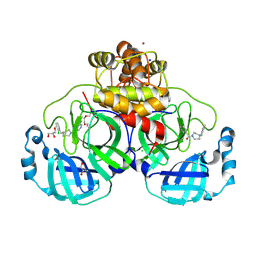 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | Descriptor: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R16
 
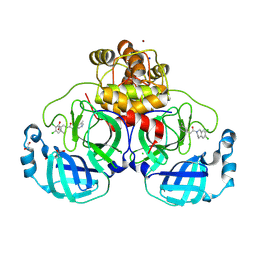 | | Structure of compound 12 bound to SARS-CoV-2 main protease | | Descriptor: | 1,2-ETHANEDIOL, 1-[6,7-bis(chloranyl)-3,4-dihydro-1H-isoquinolin-2-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
7GRE
 
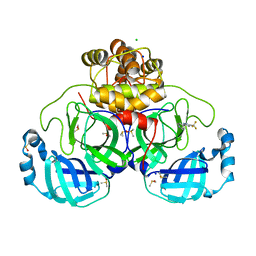 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-1 | | Descriptor: | 3C-like proteinase nsp5, 4-[3-(trifluoromethyl)-1H-pyrazol-5-yl]pyridine, CHLORIDE ION, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A.M. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6QMR
 
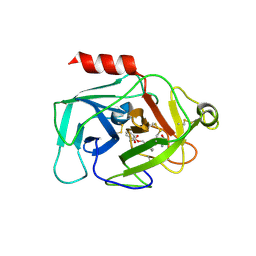 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
6QMT
 
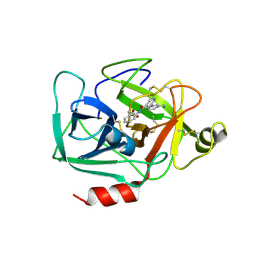 | | Complement factor D in complex with the inhibitor 2-(2-(3'-(aminomethyl)-[1,1'-biphenyl]-3-carboxamido)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
5FBE
 
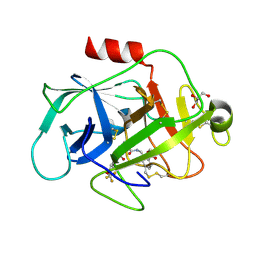 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND2 | | Descriptor: | Complement factor D, GLYCEROL, methyl 2-[[[(2~{S})-2-[[3-(trifluoromethyloxy)phenyl]carbamoyl]pyrrolidin-1-yl]carbonylamino]methyl]benzoate | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FBI
 
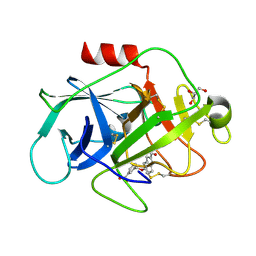 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 3b | | Descriptor: | 3-[(2-aminocarbonyl-1~{H}-indol-5-yl)oxymethyl]benzoic acid, Complement factor D, GLYCEROL | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FAH
 
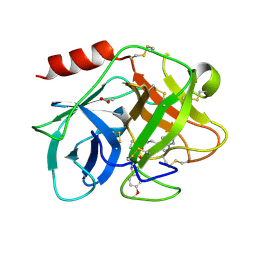 | | KALLIKREIN-7 IN COMPLEX WITH COMPOUND1 | | Descriptor: | (2~{S})-~{N}2-[2-(4-methoxyphenyl)ethyl]-~{N}1-(naphthalen-1-ylmethyl)pyrrolidine-1,2-dicarboxamide, ACETATE ION, Kallikrein-7 | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
6FTZ
 
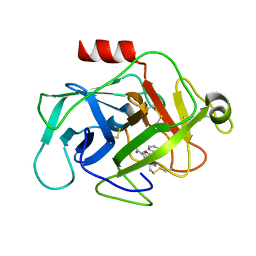 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 6 | | Descriptor: | Complement factor D, ~{N}4-[3-(aminomethyl)phenyl]-1~{H}-indole-2,4-dicarboxamide | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FTY
 
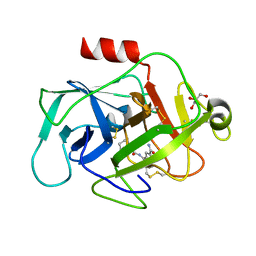 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 5 | | Descriptor: | 4-[[(5~{S},7~{R})-3-azanyl-1-adamantyl]carbonylamino]-1~{H}-indole-2-carboxamide, Complement factor D, GLYCEROL | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
9FX6
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 100K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9FX7
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 294K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
1QXZ
 
 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle inhibitor 119 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-(1,3-THIAZOL-2-YL)BUTANE-1,1-DIOL, COBALT (II) ION, methionyl aminopeptidase | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1QXW
 
 | | Crystal structure of Staphyloccocus aureus in complex with an aminoketone inhibitor 54135. | | Descriptor: | (3S)-3-AMINO-1-(CYCLOPROPYLAMINO)HEPTANE-2,2-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1QXY
 
 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle 618 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-PYRIDIN-2-YLBUTANE-1,1-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
