9HAI
 
 | | Pooled 50S subunit C-CP_L2-L28 precursor states supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Apidaecins type 22, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-04 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA1
 
 | | Pooled 50S subunit C_(L22)- precursor states supplemented with Api137 - Canonical PET exit Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Apidaecins type 137, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9FL7
 
 | |
3CF5
 
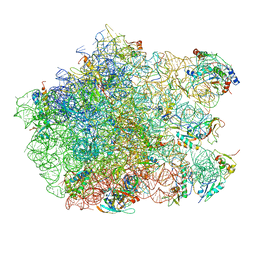 | | Thiopeptide antibiotic Thiostrepton bound to the large ribosomal subunit of Deinococcus radiodurans | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, 50S RIBOSOMAL PROTEIN L14, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Translational Regulation Via L11: Molecular Switches on the Ribosome Turned on and Off by Thiostrepton and Micrococcin.
Mol.Cell, 30, 2008
|
|
3JCN
 
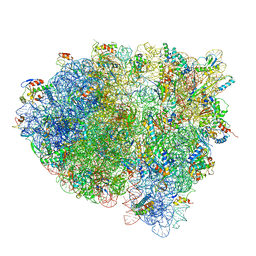 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association: Initiation Complex I | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
2OM7
 
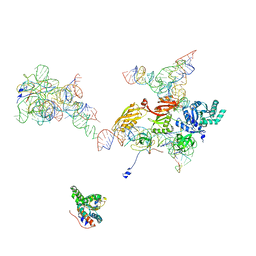 | | Structural Basis for Interaction of the Ribosome with the Switch Regions of GTP-bound Elongation Factors | | Descriptor: | 16S ribosomal RNA (H5), 30S ribosomal protein S12, 30S ribosomal protein S2, ... | | Authors: | Connell, S.R, Wilson, D.N, Rost, M, Schueler, M, Giesebrecht, J, Dabrowski, M, Mielke, T, Fucini, P, Spahn, C.M.T. | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors.
Mol.Cell, 25, 2007
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
3RGW
 
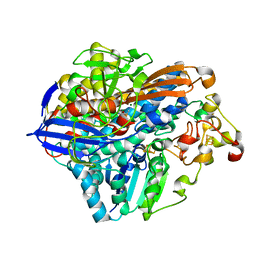 | | Crystal structure at 1.5 A resolution of an H2-reduced, O2-tolerant hydrogenase from Ralstonia eutropha unmasks a novel iron-sulfur cluster | | Descriptor: | FE3-S4 CLUSTER, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Scheerer, P, Fritsch, J, Frielingsdorf, S, Kroschinsky, S, Friedrich, B, Lenz, O, Spahn, C.M.T. | | Deposit date: | 2011-04-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre.
Nature, 479, 2011
|
|
1FCW
 
 | | TRNA POSITIONS DURING THE ELONGATION CYCLE | | Descriptor: | TRNAPHE | | Authors: | Agrawal, R.K, Spahn, C.M.T, Penczek, P, Grassucci, R.A, Nierhaus, K.H, Frank, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-08-11 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle.
J.Cell Biol., 150, 2000
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
7PIV
 
 | | Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution. | | Descriptor: | CALCIUM ION, Camelid antibody VHH fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
7SOX
 
 | |
7PIU
 
 | | Cryo-EM structure of the agonist setmelanotide bound to the active melanocortin-4 receptor (MC4R) in complex with the heterotrimeric Gs protein at 2.6 A resolution. | | Descriptor: | CALCIUM ION, Camelid antibody fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
5T4X
 
 | | CRYSTAL STRUCTURE OF PDE6D IN APO-STATE | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Qureshi, B.M, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-08-30 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanistic insights into the role of prenyl-binding protein PrBP/ delta in membrane dissociation of phosphodiesterase 6.
Nat Commun, 9, 2018
|
|
3JCJ
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
8RQ2
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation III | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RQ0
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RPY
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137 | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 137, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RPZ
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
4V6T
 
 | | Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Yamamoto, H, Rother, K, Wittek, D, Pech, M, Mielke, T, Loerke, J, Scheerer, P, Ivanov, P, Teraoka, Y, Shpanchenko, O, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The complex of tmRNA-SmpB and EF-G on translocating ribosomes.
Nature, 485, 2012
|
|
4V7B
 
 | | Visualization of two tRNAs trapped in transit during EF-G-mediated translocation | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Lancaster, L, Sprink, T, Mielke, T, Loerke, J, Noller, H.F, Spahn, C.M.T. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7ZN7
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
5M1J
 
 | | Nonstop ribosomal complex bound with Dom34 and Hbs1 | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Hilal, T, Yamamoto, H, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution.
Nat Commun, 7, 2016
|
|
2NOQ
 
 | | Structure of ribosome-bound cricket paralysis virus IRES RNA | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S5, ... | | Authors: | Schuler, M, Connell, S.R, Lescoute, A, Giesebrecht, J, Dabrowski, M, Schroeer, B, Mielke, T, Penczek, P.A, Westhof, E, Spahn, C.M.T. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure of the ribosome-bound cricket paralysis virus IRES RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
