5A78
 
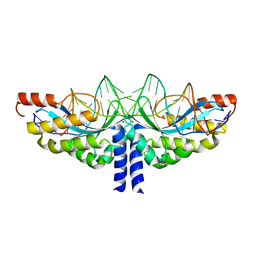 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA not cleaved | | Descriptor: | 24MER DNA, 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP *CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', DNA ENDONUCLEASE I-CVUI, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5AKN
 
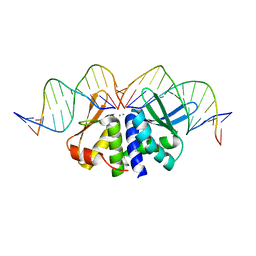 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA NICKED IN THE non-CODING STRAND B AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP *CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5A74
 
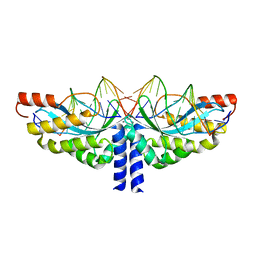 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Mn | | Descriptor: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*CP*TP*GP*AP)-3', 14MER DNA, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5AK9
 
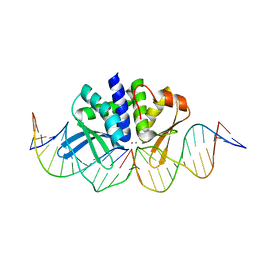 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*GP)-3, ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5AKM
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI G20S IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MG | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5A0W
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI E117A IN COMPLEX WITH ITS TARGET DNA AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, ACETATE ION, CHLORIDE ION, ... | | Authors: | Molina, R, Besker, N, Prieto, J, Montoya, G, D'Abramo, M. | | Deposit date: | 2015-04-23 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Dynamical Characterization of the I- Dmo Catalytic Activity
To be Published
|
|
5A72
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Ca | | Descriptor: | 24MER DNA, 5'-D(*DTP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*AP *DCP*GP*AP*CP*GP*TP*TP*CP*TP*GP*A)-3', CALCIUM ION, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
4AAD
 
 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in absence of metal ions at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI, GLYCEROL | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAG
 
 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in presence of Ca at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP *GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', CALCIUM ION, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AQX
 
 | | Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of magnesium | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*5CM)-3', 5'-D(*GP*AP*CP*AP*GP*TP*TP*TP*GP*GP)-3', 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', ... | | Authors: | Valton, J, Daboussi, F, Leduc, S, Redondo, P, Macmaster, R, Molina, R, Montoya, G, Duchateau, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases.
J.Biol.Chem., 287, 2012
|
|
4AAB
 
 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 10MER DNA 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', 14MER DNA 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP)-3', DNA ENDONUCLEASE I-CREI, ... | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AFL
 
 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
4AAE
 
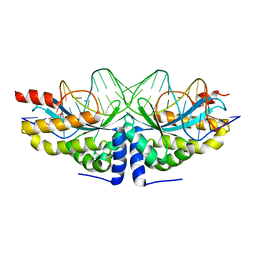 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by AGCG from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
6GTC
 
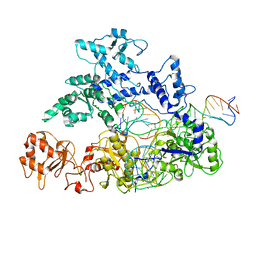 | | Transition state structure of Cpf1(Cas12a) I1 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*G)-3'), DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), ... | | Authors: | Mesa, P, Montoya, G. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
4AAF
 
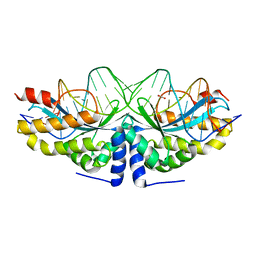 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by TGCA from 5' to 3') | | Descriptor: | 1,2-ETHANEDIOL, 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
6GTG
 
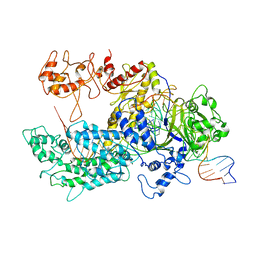 | | Transition state structure of Cpf1(Cas12a) I4 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (32-MER), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), ... | | Authors: | Mesa, P, Montoya, G, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
4AQU
 
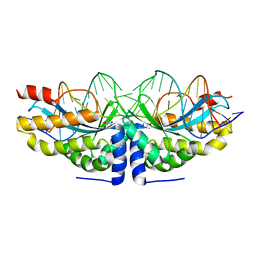 | | Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of calcium | | Descriptor: | 5'-D(*DCP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*5CMP *GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 5'-D(*DTP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*DAP *GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', CALCIUM ION, ... | | Authors: | Valton, J, Daboussi, F, Leduc, S, Redondo, P, Macmaster, R, Molina, R, Montoya, G, Duchateau, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases.
J.Biol.Chem., 287, 2012
|
|
4CJA
 
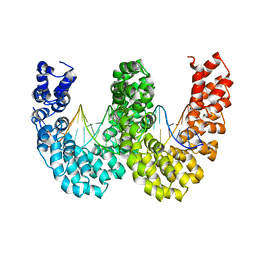 | | BurrH DNA-binding protein from Burkholderia rhizoxinica in complex with its target DNA | | Descriptor: | 5'-D(*DTP*AP*TP*AP*AP*CP*GP*TP*AP*TP*TP*TP*GP*CP *TP*TP*CP*TP*CP*TP*TP*AP*AP)-3', 5'-D(*DTP*TP*AP*AP*GP*AP*GP*AP*AP*GP*CP*AP*AP*DP *TP*AP*CP*GP*TP*TP*AP*TP*AP)-3', BURRH | | Authors: | Stella, S, Molina, R, Lopez-Mendez, B, Campos-Olivas, R, Duchateau, P, Montoya, G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6QZT
 
 | |
6R7B
 
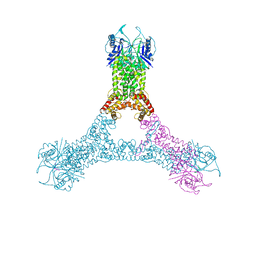 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 1 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6R9R
 
 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 2 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, circular RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6QZQ
 
 | |
8QBM
 
 | |
8QBL
 
 | |
8QBK
 
 | |
