3ZNZ
 
 | | Crystal structure of OTULIN OTU domain (C129A) in complex with Met1- di ubiquitin | | Descriptor: | POLYUBIQUITIN-C, PROTEIN FAM105B, SULFATE ION | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZNV
 
 | | Crystal structure of the OTU domain of OTULIN at 1.3 Angstroms. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
7MJI
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJJ
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJL
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJH
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJK
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
8CRO
 
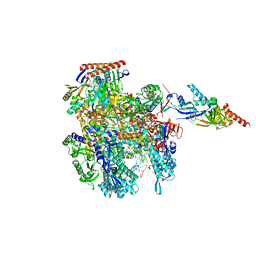 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
3LPE
 
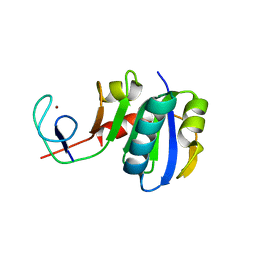 | | Crystal structure of Spt4/5NGN heterodimer complex from Methanococcus jannaschii | | Descriptor: | DNA-directed RNA polymerase subunit E'', Putative transcription antitermination protein nusG, ZINC ION | | Authors: | Hirtreiter, A, Damsma, G.E, Cheung, A.C.M, Klose, D, Grohmann, D, Vojnic, E, Martin, A.C.R, Cramer, P, Werner, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif.
Nucleic Acids Res., 38, 2010
|
|
5AFW
 
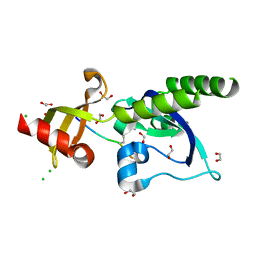 | | Assembly of methylated LSD1 and CHD1 drives AR-dependent transcription and translocation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1, ... | | Authors: | Metzger, E, Willmann, D, McMillan, J, Petroll, K, Metzger, P, Gerhardt, S, vonMaessenhausen, A, Schott, A.K, Espejo, A, Eberlin, A, Wohlwend, D, Schuele, K.M, Schleicher, M, Perner, S, Bedford, M.T, Dengjel, J, Flaig, R, Einsle, O, Schuele, R. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3EG5
 
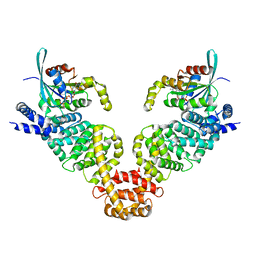 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
1PLG
 
 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
2PPZ
 
 | |
1NQ7
 
 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | 7-(3,5-DITERT-BUTYLPHENYL)-3-METHYLOCTA-2,4,6-TRIENOIC ACID, NUCLEAR RECEPTOR ROR-BETA, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
1N4H
 
 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | Nuclear Receptor ROR-beta, RETINOIC ACID, Steroid Receptor Coactivator-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2002-10-31 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
2B5O
 
 | | ferredoxin-NADP reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, SULFATE ION | | Authors: | Sawaya, M.R, Kerfeld, C.A, Gomez-Lojero, C, Krogmann, D, Bryant, D.A, Yeates, T.O. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Crystal Structure of Ferredoxin-NADP reductase from Synechococcus sp. (PCC 7002)
To be Published
|
|
1JUT
 
 | | E. coli Thymidylate Synthase Bound to dUMP and LY338529, A Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-3-METHYL-BUTYRIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1JUJ
 
 | | Human Thymidylate Synthase Bound to dUMP and LY231514, a Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1KQE
 
 | | Solution structure of a linked shortened gramicidin A in benzene/acetone 10:1 | | Descriptor: | MINI-GRAMICIDIN A | | Authors: | Arndt, H.D, Bockelmann, D, Knoll, A, Lamberth, S, Griesinger, C, Koert, U. | | Deposit date: | 2002-01-05 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Cation Control in Functional Helical Programming: Structures of a D,L-Peptide Ion Channel
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
1JTQ
 
 | | E. coli TS Complex with dUMP and the Pyrrolo(2,3-d)pyrimidine-based Antifolate LY341770 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-4-(2H-TETRAZOL-5-YL)-BUTYRIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-21 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1JU6
 
 | | Human Thymidylate Synthase Complex with dUMP and LY231514, A Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, PHOSPHATE ION, ... | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1JTU
 
 | | E. coli Thymidylate Synthase in a Complex with dUMP and LY338913, A Polyglutamylated Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[4-(4-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-4-CARBOXY-BUTYRYLAMIN O)-4-CARBOXY-BUTYRYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-22 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
