6ZLM
 
 | | Dihydrolipoyllysine-residue acetyltransferase component of fungal pyruvate dehydrogenase complex with protein X bound | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, Pyruvate dehydrogenase X component | | Authors: | Forsberg, B.O, Aibara, S, Howard, R.J, Mortezaei, N, Lindahl, E. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
8QCB
 
 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, Superkiller protein 3, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8QCA
 
 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the closed state bound to RNA | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8Q9T
 
 | | CryoEM structure of a S. Cerevisiae Ski238 complex bound to RNA | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8QCF
 
 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
6ZLO
 
 | | E2 core of the fungal Pyruvate dehydrogenase complex with asymmetric interior PX30 component | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Forsberg, B.O, Howard, R.J, Aibara, S, Mortesaei, N, Lindahl, E. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
3HO0
 
 | | Crystal structure of the PPARgamma-LBD complexed with a new aryloxy-3phenylpropanoic acid | | Descriptor: | (2S)-2-(4-phenethylphenoxy)-3-phenyl-propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Laghezza, A, Lavecchia, A, Novellino, E. | | Deposit date: | 2009-06-01 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New 2-Aryloxy-3-phenyl-propanoic Acids As Peroxisome Proliferator-Activated Receptors alpha/gamma Dual Agonists with Improved Potency and Reduced Adverse Effects on Skeletal Muscle Function
J.Med.Chem., 52, 2009
|
|
5LFF
 
 | | NMR structure of peptide 2 targeting CXCR4 | | Descriptor: | ARG-ALA-CYS-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
3HOD
 
 | | Crystal structure of the PPARgamma-LBD complexed with a new aryloxy-3phenylpropanoic acid | | Descriptor: | (2S)-2-(4-benzylphenoxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Laghezza, A, Lavecchia, A, Novellino, E. | | Deposit date: | 2009-06-02 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New 2-Aryloxy-3-phenyl-propanoic Acids As Peroxisome Proliferator-Activated Receptors alpha/gamma Dual Agonists with Improved Potency and Reduced Adverse Effects on Skeletal Muscle Function
J.Med.Chem., 52, 2009
|
|
6HUE
 
 | | ParkinS65N | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | McWilliams, T.G, Barini, E, Pohjolan-Pirhonen, R, Brooks, S.P, Singh, F, Burel, S, Balk, K, Kumar, A, Montava-Garriga, L, Prescott, A.R, Hassoun, S.M, Mouton-Liger, F, Ball, G, Hills, R, Knebel, A, Ulusoy, A, Di Monte, D.A, Tamjar, J, Antico, O, Fears, K, Smith, L, Brambilla, R, Palin, E, Valori, M, Eerola-Rautio, J, Tienari, P, Corti, O, Dunnett, S.B, Ganley, I.G, Suomalainen, A, Muqit, M.M.K. | | Deposit date: | 2018-10-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Phosphorylation of Parkin at serine 65 is essential for its activation in vivo .
Open Biology, 8, 2018
|
|
6HVK
 
 | | Pepducin UT-Pep2 a biased allosteric agonist of Urotensin-II receptor | | Descriptor: | Urotensin-2 receptor | | Authors: | Carotenuto, A, Hoang, T.A, Nassour, H, Martin, R.D, Billard, E, Myriam, L, Novellino, E, Tanny, J.C, Fournier, A, Hebert, T.E, Chatenet, D. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lipidated peptides derived from intracellular loops 2 and 3 of the urotensin II receptor act as biased allosteric ligands.
J.Biol.Chem., 297, 2021
|
|
8OQ6
 
 | | CryoEM structure of human rho1 GABAA receptor apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQA
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQ7
 
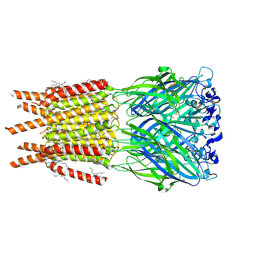 | | CryoEM structure of human rho1 GABAA receptor in complex with inhibitor TPMPA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQ8
 
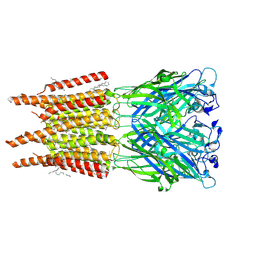 | | CryoEM structure of human rho1 GABAA receptor in complex with pore blocker picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OP9
 
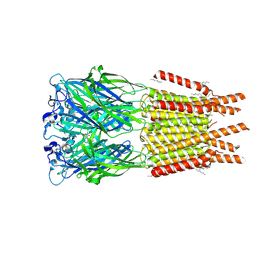 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OWY
 
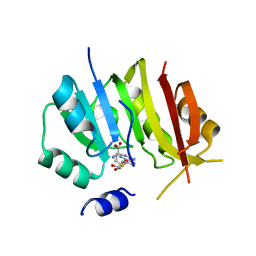 | | Crystal structure of METTL6 mutant 40-269 bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8P7D
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 1:1:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8P7B
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8P7C
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 2:2:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7Q3H
 
 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM EDTA | | Descriptor: | Neur_chan_LBD domain-containing protein | | Authors: | Lycksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q3G
 
 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM Ca2+ | | Descriptor: | CALCIUM ION, Neur_chan_LBD domain-containing protein | | Authors: | Licksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QWS
 
 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
7QWQ
 
 | | Ternary complex of ribosome nascent chain with SRP and NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
