1GHQ
 
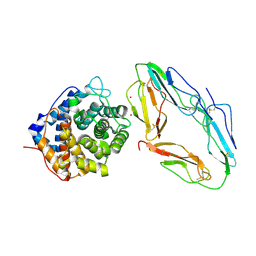 | | CR2-C3D COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, COMPLEMENT C3, CR2/CD121/C3D/EPSTEIN-BARR VIRUS RECEPTOR, ... | | Authors: | Szakonyi, G, Guthridge, J.M, Li, D, Holers, V.M, Chen, X.S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of complement receptor 2 in complex with its C3d ligand.
Science, 292, 2001
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
5XSG
 
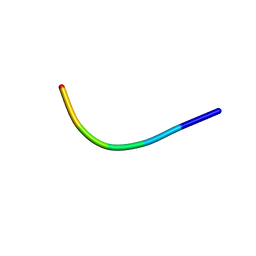 | | Ultrahigh resolution structure of FUS (37-42) SYSGYS determined by MicroED | | Descriptor: | RNA-binding protein FUS | | Authors: | Luo, F, Gui, X, Zhou, H, Li, D, Li, X, Liu, C. | | Deposit date: | 2017-06-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.73 Å) | | Cite: | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8ZMO
 
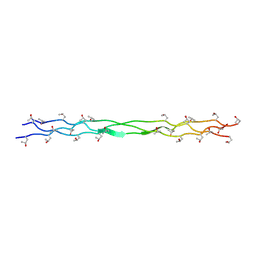 | | Structure of a triple-helix region of human Collagen type XVII from Trautec | | Descriptor: | collagen type XVII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type XVII from Trautec
To Be Published
|
|
8ZMW
 
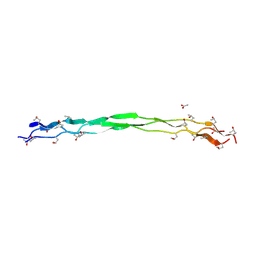 | | Structure of a triple-helix region of human Collagen type VII from Trautec | | Descriptor: | ACETATE ION, collagen type VII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a triple-helix region of human Collagen type VII from Trautec
To Be Published
|
|
8ZMM
 
 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | collagen type IV | | Authors: | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
1I1H
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PRECORRIN-8X METHYLMUTASE COMPLEX WITH HYDROGENOBYRINIC ACID | | Descriptor: | HYDROGENOBYRINIC ACID, PRECORRIN-8X METHYLMUTASE | | Authors: | Shipman, L.W, Li, D, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-02-01 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of precorrin-8x methyl mutase.
Structure, 9, 2001
|
|
8Q8S
 
 | | Tau - AD-THF | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q8Z
 
 | |
8Q9A
 
 | |
8Q8E
 
 | |
8Q8X
 
 | |
8Q9B
 
 | |
6A6B
 
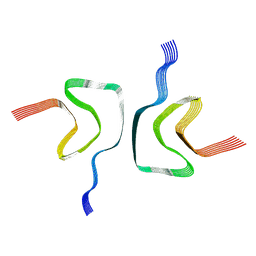 | | cryo-em structure of alpha-synuclein fiber | | Descriptor: | Alpha-synuclein | | Authors: | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | Deposit date: | 2018-06-27 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
9IJP
 
 | |
8K9R
 
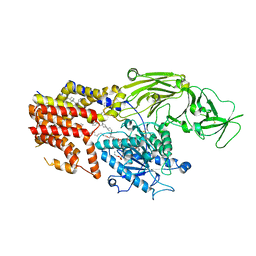 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9Q
 
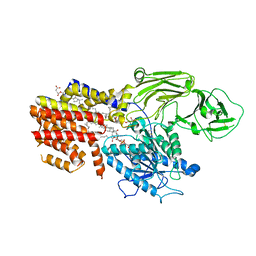 | | Cryo-EM structure of the GPI inositol-deacylase (PGAP1/Bst1) from Chaetomium thermophilum | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, CHOLESTEROL HEMISUCCINATE, GPI inositol-deacylase,fused thermostable green fluorescent protein | | Authors: | Hong, J, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9T
 
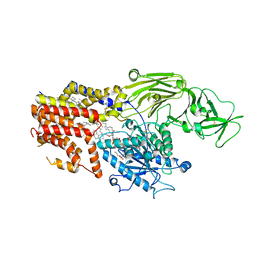 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7F5G
 
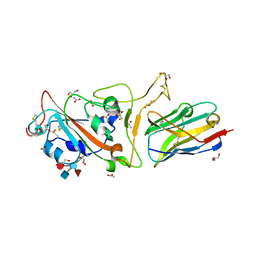 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | Descriptor: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | Authors: | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
