8CTN
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction, no electric field) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTT
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) at 100K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTW
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Na+,Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTX
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -K+,Tl+ complex | | Descriptor: | POTASSIUM ION, Potassium channel protein, THALLIUM (I) ION | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTV
 
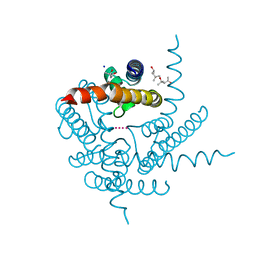 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTS
 
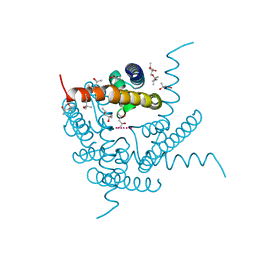 | | Room temperature crystal structure of a K+ selective NaK mutant (NaK2K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTU
 
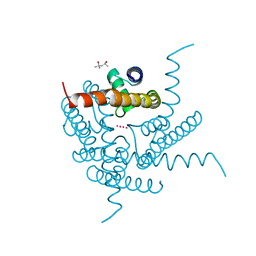 | | Crystal structure of a K+ selective NaK mutant (NaK2K) at Room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU1
 
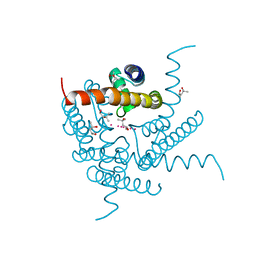 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 500ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU3
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 200ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU4
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 1us, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU2
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 100ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
1ZYI
 
 | | Solution structure of ICLN, a multifunctional protein involved in regulatory mechanisms as different as cell volume regulation and rna splicing | | Descriptor: | Methylosome subunit pICln | | Authors: | Fuerst, J, Schedlbauer, A, Gandini, R, Garavaglia, M.L, Siano, S, Gschwentner, M, Sarg, B, Kontaxis, G, Konrat, R, Paulmichl, M. | | Deposit date: | 2005-06-10 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ICln159 folds into a pleckstrin homology domain-like structure. Interaction with kinases and the splicing factor LSm4
J.Biol.Chem., 280, 2005
|
|
8R61
 
 | |
7S4Z
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Proteinase K | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4W
 
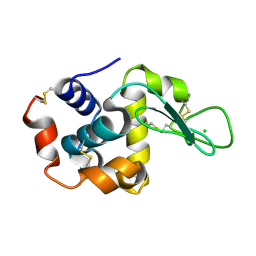 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4Y
 
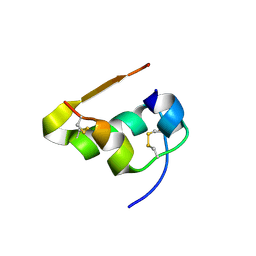 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S50
 
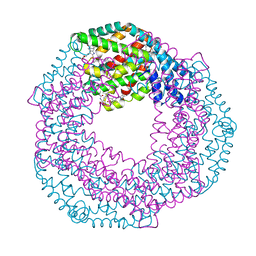 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Phycocyanin | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, CHLORIDE ION, ... | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4R
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - alpha Spectrin-SH3 domain | | Descriptor: | Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
5UZE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P182
To Be Published
|
|
5UZC
 
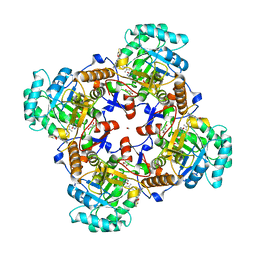 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P221 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P221
To Be Published
|
|
5UPV
 
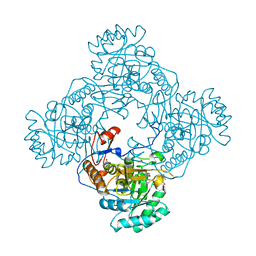 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36
To Be Published
|
|
5UTX
 
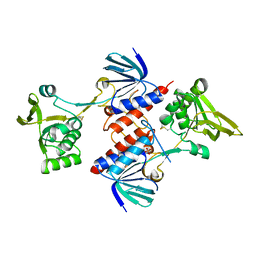 | | Crystal structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 - apo form | | Descriptor: | PHOSPHATE ION, Thioredoxin reductase | | Authors: | Chang, C, Grimshaw, S, Maltseva, N, Mulligan, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 - apo form
To Be Published
|
|
5V0I
 
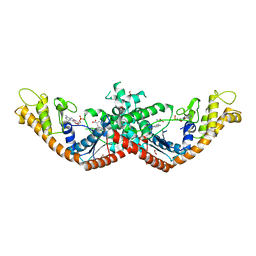 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, TRYPTOPHAN, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Grimshaw, S.G, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan
To Be Published
|
|
5VSV
 
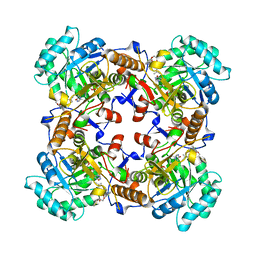 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P225 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, {2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenoxy}acetic acid | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P225
To Be Published
|
|
5VT3
 
 | | High resolution structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 in complex with NADP and FAD | | Descriptor: | CACODYLATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Chang, C, Grimshaw, S, Maltseva, N, Mulligan, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High resolution structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 in complex with NADP and FAD
To Be Published
|
|
