4B2G
 
 | | Crystal Structure of an Indole-3-Acetic Acid Amido Synthase from Vitis vinifera Involved in Auxin Homeostasis | | Descriptor: | GH3-1 AUXIN CONJUGATING ENZYME, MALONATE ION, [(2S,3R,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl] 2-(1H-indol-3-yl)ethyl hydrogen phosphate | | Authors: | Peat, T.S, Bottcher, C, Newman, J, Lucent, D, Cowieson, N, Davies, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of an Indole-3-Acetic Acid Amido Synthetase from Grapevine Involved in Auxin Homeostasis.
Plant Cell, 24, 2012
|
|
8ABX
 
 | | Crystal structure of IDO1 in complex with Apoxidole-1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, O1-tert-butyl O2-ethyl O5-methyl (E,5R)-5-(1-methylindol-2-yl)-5-[(4-methylphenyl)sulfonylamino]pent-2-ene-1,2,5-tricarboxylate, O2-tert-butyl O3-ethyl O6-methyl (2S,6R)-6-(1-methylindol-2-yl)-2,5-dihydro-1H-pyridine-2,3,6-tricarboxylate, ... | | Authors: | Dotsch, L, Ziegler, S, Waldmann, H, Gasper, R. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Pseudo-Natural Product Type IV IDO1 Inhibitor Chemotype.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
9MDZ
 
 | | anti-IL23R VHH | | Descriptor: | GLYCEROL, anti-IL23R VHH | | Authors: | Kiefer, J.R, Koerber, J.T, Ota, N, Davies, C, Wallweber, H.A. | | Deposit date: | 2024-12-05 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Engineering a protease-stable, oral single-domain antibody to inhibit IL-23 signaling.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9MFG
 
 | | Complex of IL23 receptor and VHH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Wallweber, H.A, Koerber, J.T, Ota, N, Davies, C. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Engineering a protease-stable, oral single-domain antibody to inhibit IL-23 signaling.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5AAU
 
 | | Optimization of a novel binding motif to to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H- pyrido(3,4-b)indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist | | Descriptor: | 3-(1-(4-Chlorophenyl)-3,4-dihydro-1H-pyrido(3,4-b)indol-2(9H)-yl)propanoic acid, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Bradbury, R.H, de Almeida, C, Andrews, D.M, Ballard, P, Buttar, D, Callis, R.J, Currie, G.S, Curwen, J.O, Davies, C.D, de Savi, C, Donald, C.S, Feron, L.J.L, Glossop, S.C, Hayter, B.R, Karoutchi, G, Lamont, S.G, MacFaul, P, Moss, T, Pearson, S.E, Rabow, A.A, Tonge, M, Walker, G.E, Weir, H.M, Wilson, Z. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
5AAV
 
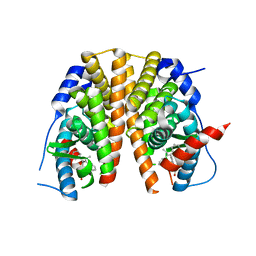 | | Optimization of a novel binding motif to to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H- pyrido(3,4-b)indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Bradbury, R.H, de Almeida, C, Andrews, D.M, Ballard, P, Buttar, D, Callis, R.J, Currie, G.S, Curwen, J.O, Davies, C.D, de Savi, C, Donald, C.S, Feron, L.J.L, Glossop, S.C, Hayter, B.R, Karoutchi, G, Lamont, S.G, MacFaul, P, Moss, T, Pearson, S.E, Rabow, A.A, Tonge, M, Walker, G.E, Weir, H.M, Wilson, Z. | | Deposit date: | 2015-07-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
7UVF
 
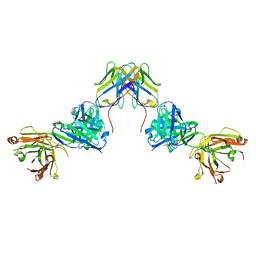 | | Crystal structure of ZED8 Fab complex with CD8 alpha | | Descriptor: | CHLORIDE ION, GLYCEROL, Immunoglobulin heavy chain, ... | | Authors: | Yu, C, Davies, C, Koerber, J.T, Williams, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Preclinical development of ZED8, an 89 Zr immuno-PET reagent for monitoring tumor CD8 status in patients undergoing cancer immunotherapy.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
2VGK
 
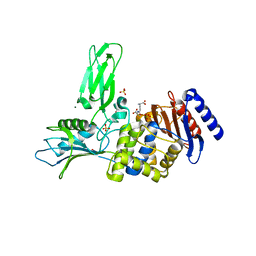 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
1QD7
 
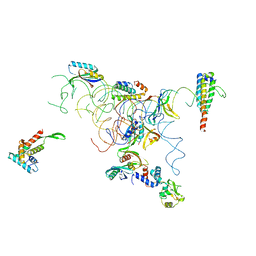 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
3Q2G
 
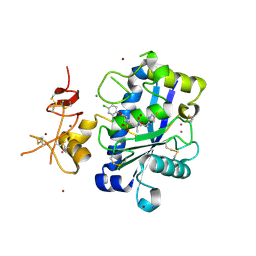 | | Adamts1 in complex with a novel N-hydroxyformamide inhibitors | | Descriptor: | A disintegrin and metalloproteinase with thrombospondin motifs 1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The design and synthesis of novel N-hydroxyformamide inhibitors of ADAM-TS4 for the treatment of osteoarthritis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q2H
 
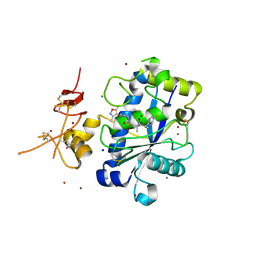 | | Adamts1 in complex with N-hydroxyformamide inhibitors of ADAM-TS4 | | Descriptor: | A disintegrin and metalloproteinase with thrombospondin motifs 1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The design and synthesis of novel N-hydroxyformamide inhibitors of ADAM-TS4 for the treatment of osteoarthritis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7SOL
 
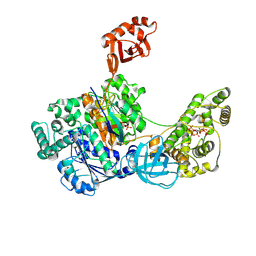 | | Crystal Structures of the bispecific ubiquitin/FAT10 activating enzyme, Uba6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Ubiquitin, ... | | Authors: | Olsen, S.K, Gao, F, Lv, Z. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25000644 Å) | | Cite: | Crystal structures reveal catalytic and regulatory mechanisms of the dual-specificity ubiquitin/FAT10 E1 enzyme Uba6.
Nat Commun, 13, 2022
|
|
8QZD
 
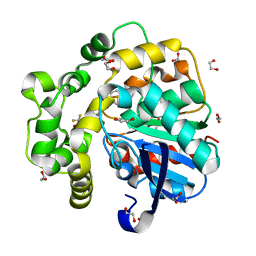 | | Soluble epoxide hydrolase in complex with Epoxykinin | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-bromanyl-3-[2,2,2-tris(fluoranyl)ethanoyl]indol-1-yl]-N-cycloheptyl-ethanamide, BROMIDE ION, ... | | Authors: | Kumar, A, Ehrler, J.M.H, Ziegler, S, Doetsch, L, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the sEH Inhibitor Epoxykynin as a Potent Kynurenine Pathway Modulator.
J.Med.Chem., 67, 2024
|
|
6CWZ
 
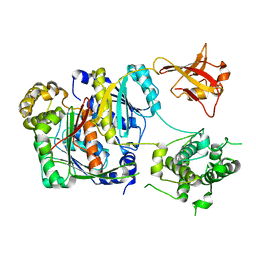 | | Crystal structure of apo SUMO E1 | | Descriptor: | SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ZINC ION | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
6CWY
 
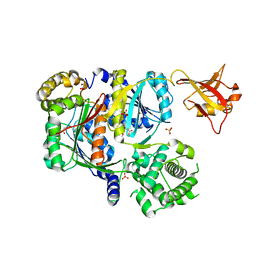 | | Crystal structure of SUMO E1 in complex with an allosteric inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
6BR7
 
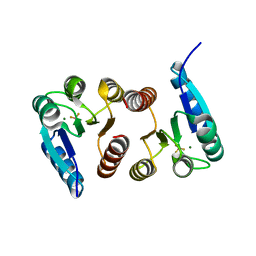 | |
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
2VGJ
 
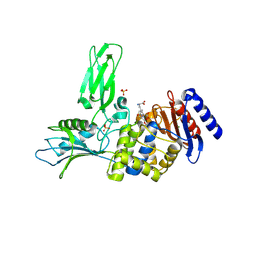 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | CEPHALOSPORIN, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
2NAZ
 
 | | The solution NMR structure of the C-terminal effector domain of BfmR from Acinetobacter baumannii | | Descriptor: | Transcriptional regulatory protein RstA | | Authors: | Olson, A.L, Thompson, R.J, Cavanagh, J, Feldmann, E.A, Bobay, B.G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Biofilm-controlling Response Regulator BfmR from Acinetobacter baumannii Reveals Details of Its DNA-binding Mechanism.
J.Mol.Biol., 430, 2018
|
|
1XF3
 
 | | Structure of ligand-free Fab DNA-1 in space group P65 | | Descriptor: | Fab Light chain, Fab heavy chain | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
1XF4
 
 | | Structure of ligand-free Fab DNA-1 in space group P321 solved from crystals with perfect hemihedral twinning | | Descriptor: | Fab heavy chain, Fab light chain, SULFATE ION | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
487D
 
 | |
1XF2
 
 | | Structure of Fab DNA-1 complexed with dT3 | | Descriptor: | 5'-D(*TP*TP*T)-3', SULFATE ION, antibody heavy chain Fab, ... | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
1CQU
 
 | |
1FKA
 
 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
