4FD2
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FCV
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FCT
 
 | |
4FCW
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4NM1
 
 | | Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-C at N-1 position and G-C at N position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*(BGM)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4NM2
 
 | | Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-G at N-1 position and G-C at N position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*(BGM)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.524 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
6E3R
 
 | |
6E3V
 
 | |
6E3W
 
 | |
6E3X
 
 | |
7TFM
 
 | | Atomic Structure of the Leishmania spp. Hsp100 N-Domain | | Descriptor: | ATP-dependent Clp protease subunit, heat shock protein 100 (HSP100), GLYCEROL | | Authors: | Mercado, J.M, Lee, S, Chang, C, Sung, N, Soong, L, Catic, A, Tsai, F.T.F. | | Deposit date: | 2022-01-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.055 Å) | | Cite: | Atomic structure of the Leishmania spp. Hsp100 N-domain.
Proteins, 90, 2022
|
|
8GW7
 
 | | AtSLAC1 6D mutant in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8GW6
 
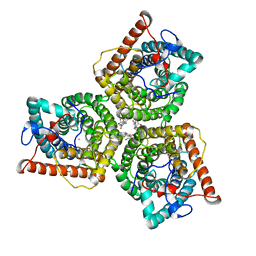 | | AtSLAC1 6D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J1E
 
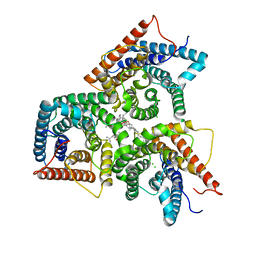 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
2SNW
 
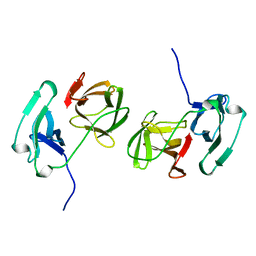 | | SINDBIS VIRUS CAPSID PROTEIN, TYPE3 CRYSTAL FORM | | Descriptor: | COAT PROTEIN C | | Authors: | Choi, H.-K, Lee, S, Zhang, Y.-P, Mckinney, B.R, Wengler, G, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 1998-02-17 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis.
J.Mol.Biol., 262, 1996
|
|
4TUR
 
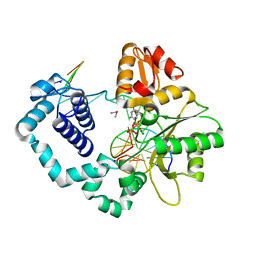 | |
4TUS
 
 | |
4TUQ
 
 | | Human DNA polymerase beta inserting dCMPNPP opposite GG template (GG0b). | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*CP*CP*AP*CP*GP*GP*CP*CP*CP*AP*TP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*AP*TP*GP*GP*GP*C)-3'), ... | | Authors: | Koag, M.C, Lee, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Structural Basis for the Inefficient Nucleotide Incorporation Opposite Cisplatin-DNA Lesion by Human DNA Polymerase beta.
J.Biol.Chem., 289, 2014
|
|
4TUP
 
 | |
5GWO
 
 | | Crystal structure of RCAR3:PP2C S265F/I267M with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
5GWP
 
 | | Crystal structure of RCAR3:PP2C wild-type with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
5ZCU
 
 | | Crystal structure of RCAR3:PP2C wild-type with pyrabactin | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, Y, Lee, S. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural determinants for pyrabactin recognition in ABA receptors in Oryza sativa.
Plant Mol.Biol., 100, 2019
|
|
7N1J
 
 | |
7N1K
 
 | |
