5X8F
 
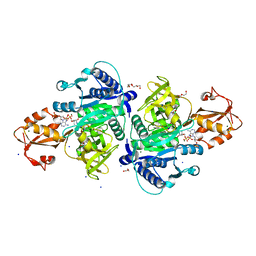 | |
8WKN
 
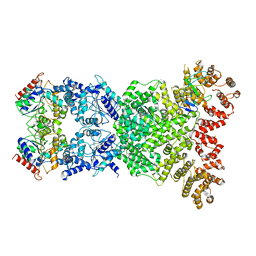 | | Cryo-EM structure of DSR2-DSAD1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
5X8G
 
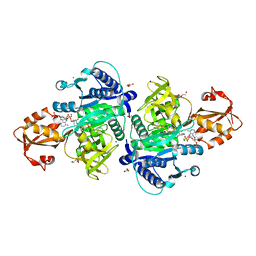 | |
7BRC
 
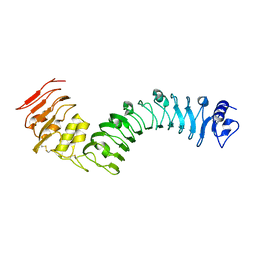 | | Crystal structure of the TMK3 LRR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-like kinase TMK3 | | Authors: | Chen, H, Kong, Y.Q, Chen, J, Li, L, Li, X.S, Yu, F, Ming, Z.H. | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the extracellular domain of the receptor-like kinase TMK3 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | Descriptor: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | Descriptor: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | Authors: | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5V7U
 
 | |
3E3R
 
 | |
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
8S9M
 
 | | DNA cytosine-N4 methyltransferase (residues 79-324) from the Bdelloid rotifer Adineta vaga | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9O
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9N
 
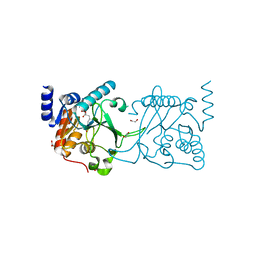 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8SDG
 
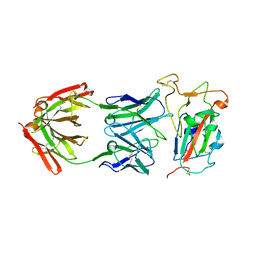 | |
8I88
 
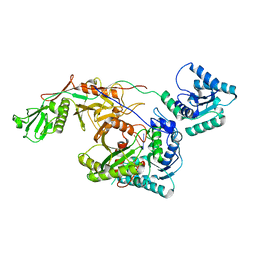 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | Descriptor: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
4O1V
 
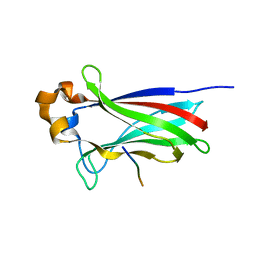 | | SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, Speckle-type POZ protein | | Authors: | Calabrese, M.F, Watson, E.R, Schulman, B.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer.
Cancer Cell, 25, 2014
|
|
4IF2
 
 | |
1W66
 
 | |
6IRO
 
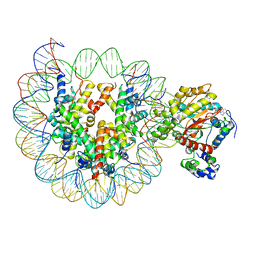 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2018-11-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
8DYP
 
 | |
6A7A
 
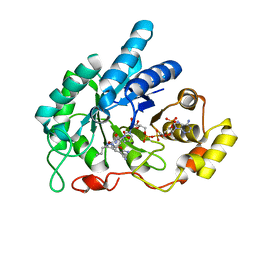 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6A7B
 
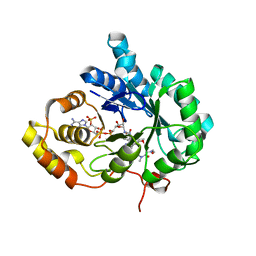 | | AKR1C3 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
4JWF
 
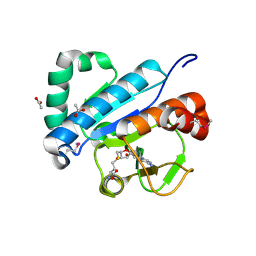 | | Crystal structure of spTrm10(74)-SAH complex | | Descriptor: | ACETIC ACID, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine(9)-N1)-methyltransferase | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2013-03-27 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
9I4H
 
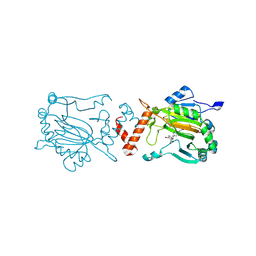 | | Factor Inhibiting HIF (FIH) in complex with manganese and 3-Hydroxy-5-(3-(4-(hydroxymethyl)-3-nitrophenyl)isoxazol-5-yl)picolinoyl)glycine | | Descriptor: | 2-[[5-[3-[4-(hydroxymethyl)-3-nitro-phenyl]-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, MANGANESE (II) ION | | Authors: | Kaur, S, Zhang, X.J, Schofield, C.J. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-Induced, Lysine-Targeting Irreversible Covalent Inhibition of the Human Oxygen Sensing Hydroxylase Factor Inhibiting HIF (FIH).
J.Am.Chem.Soc., 147, 2025
|
|
4JR6
 
 | | Crystal structure of DsbA from Mycobacterium tuberculosis (reduced) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
7F3B
 
 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
