3DL7
 
 | | Aged Form of Mouse Acetylcholinesterase Inhibited by Tabun- Update | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Carletti, E, Li, H, Li, B, Ekstrom, F, Nicolet, Y, Loiodice, M, Gillon, E, Froment, M.T, Lockridge, O, Schopfer, L.M, Masson, P, Nachon, F. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
7RD6
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD8
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E1-ATP state | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.64 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD7
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P-transition state | | Descriptor: | MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1, TETRAFLUOROALUMINATE ION | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
8GAO
 
 | | bacteriophage T4 stalled primosome with mutant gp41-E227Q | | Descriptor: | DNA (70-mer), DNA primase, DnaB-like replicative helicase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8G0Z
 
 | |
8E0Q
 
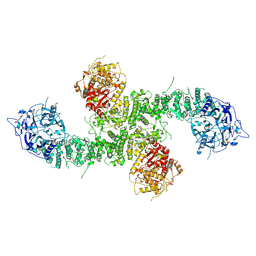 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a C2 symmetric dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
7RQH
 
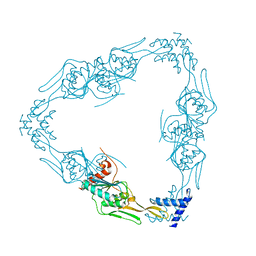 | |
7RQF
 
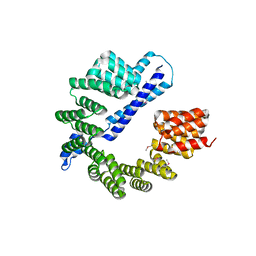 | |
5H3O
 
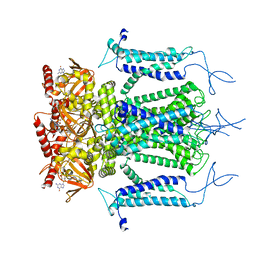 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
7RPQ
 
 | |
1FOJ
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 7-NITROINDAZOLE-2-CARBOXAMIDINE (H4B PRESENT) | | Descriptor: | 7-NITROINDAZOLE, 7-NITROINDAZOLE-2-CARBOXAMIDINE, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism
Biochemistry, 40, 2001
|
|
3DL4
 
 | | Non-Aged Form of Mouse Acetylcholinesterase Inhibited by Tabun- Update | | Descriptor: | Acetylcholinesterase, HEXAETHYLENE GLYCOL | | Authors: | Carletti, E, Li, H, Li, B, Ekstrom, F, Nicolet, Y, Loiodice, M, Gillon, E, Froment, M.T, Lockridge, O, Schopfer, L.M, Masson, P, Nachon, F. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
8D2N
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Pre-cleavage) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2P
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA target strand (5'-D(P*CP*CP*AP*GP*GP*AP*TP*CP*TP*TP*GP*CP*CP*AP*TP*CP*CP*TP*AP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2K
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 2) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*GP*A)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2O
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2L
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 1) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*GP*A)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2Q
 
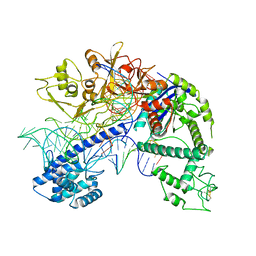 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 1) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D6Y
 
 | |
8D6W
 
 | |
8D6V
 
 | |
8D6X
 
 | |
5BKH
 
 | |
8EWI
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a tetrameric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-10-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
