5XOS
 
 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
8PVQ
 
 | | Crystal Structure of Human PTX3 C-terminal domain | | Descriptor: | Pentraxin-related protein PTX3 | | Authors: | Levy, C.W. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Pentraxin 3 mediated crosslinking of heavy chain hyaluronan complexes: new insights from structural and biophysical studies
To Be Published
|
|
5JHL
 
 | | Crystal structure of zika virus envelope protein in complex with a flavivirus broadly-protective antibody | | Descriptor: | Antibody Heavy chain, antibody Light chain, envelope protein | | Authors: | Dai, L, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Cell Host Microbe, 19, 2016
|
|
4FA6
 
 | |
6NFX
 
 | | MBTD1 MBT repeats | | Descriptor: | GLYCEROL, MBT domain-containing protein 1,Enhancer of polycomb homolog 1, SODIUM ION, ... | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-21 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex.
Cell Rep, 30, 2020
|
|
4FAD
 
 | | Design and Synthesis of a Novel Pyrrolidinyl Pyrido Pyrimidinone Derivative as a Potent Inhibitor of PI3Ka and mTOR | | Descriptor: | 2-amino-6-(5-fluoro-6-methoxypyridin-3-yl)-4-methyl-8-(pyrrolidin-1-yl)pyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Greasley, S.E, Knighton, D.R, LaFleur Rogers, C.M. | | Deposit date: | 2012-05-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and synthesis of a novel pyrrolidinyl pyrido pyrimidinone derivative as a potent inhibitor of PI3Kalpha and mTOR
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8HWL
 
 | | Human Pyruvate Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (5.63 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 2024
|
|
3DBN
 
 | | Crystal structure of the Streptoccocus suis serotype2 D-mannonate dehydratase in complex with its substrate | | Descriptor: | D-MANNONIC ACID, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q, Gao, F, Gao, G.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
4AOI
 
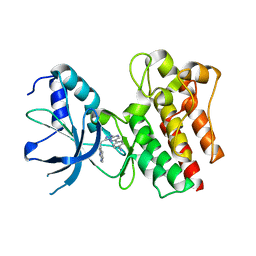 | | Crystal structure of C-MET kinase domain in complex with 4-(3-((1H- pyrrolo(2,3-b)pyridin-3-yl)methyl)-(1,2,4)triazolo(4,3-b)(1,2,4) triazin-6-yl)benzonitrile | | Descriptor: | 4-[3-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-6-yl]benzenecarbonitrile, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
4AP7
 
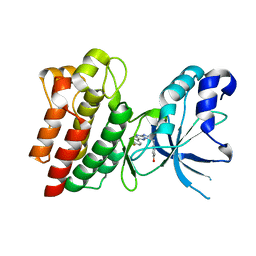 | | Crystal structure of C-MET kinase domain in complex with 4-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl)phenol | | Descriptor: | 4-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]phenol, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
3FVM
 
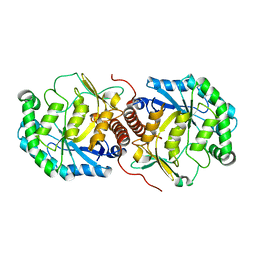 | | Crystal structure of Steptococcus suis mannonate dehydratase with metal Mn++ | | Descriptor: | MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q.J, Gao, F, Liu, Y, Gao, F.G. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
3ZXZ
 
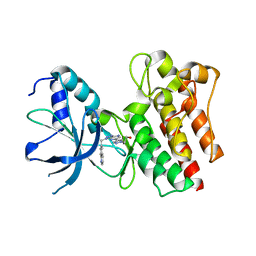 | | X-ray Structure of PF-04217903 bound to the kinase domain of c-Met | | Descriptor: | 2-{4-[1-(QUINOLIN-6-YLMETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRAZIN-6-YL]-1H-PYRAZOL-1-YL}ETHANOL, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-16 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
3ZZE
 
 | | Crystal structure of C-MET kinase domain in complex with N'-((3Z)-4- chloro-7-methyl-2-oxo-1,2-dihydro-3H-indol-3-ylidene)-2-(4- hydroxyphenyl)propanohydrazide | | Descriptor: | (2S)-N'-[(3R)-4-chloro-7-methyl-2-oxo-2,3-dihydro-1H-indol-3-yl]-2-(4-hydroxyphenyl)propanehydrazide, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Deng, Y, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-31 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
8GTK
 
 | |
8GTJ
 
 | |
6AEE
 
 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | Descriptor: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Wang, Q, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2018-08-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
6AED
 
 | |
6IEW
 
 | |
6IW2
 
 | | Crystal structure of 5A ScFv in complex with YFV-17D sE in prefusion state | | Descriptor: | Envelope protein E, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | Authors: | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | Deposit date: | 2018-12-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
5UG9
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UGA
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium | | Descriptor: | 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
6IW5
 
 | |
5UGB
 
 | |
6IW4
 
 | |
4EN0
 
 | | Crystal structure of light | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhan, C, Liu, W, Patskovsky, Y, Ramagopal, U.A, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
