6KWO
 
 | | Crystal structure of pSLA-1*1301 complex with mutant epitope ESDTVGWSW | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
6E24
 
 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5U81
 
 | | Acid ceramidase (ASAH1, aCDase) from naked mole rat, Cys143Ala, uncleaved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid ceramidase isoform b, CHLORIDE ION, ... | | Authors: | Gebai, A, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the activation of acid ceramidase.
Nat Commun, 9, 2018
|
|
7EW7
 
 | | Cryo-EM structure of SEW2871-bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 5-[4-phenyl-5-(trifluoromethyl)thiophen-2-yl]-3-[3-(trifluoromethyl)phenyl]-1,2,4-oxadiazole, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
6KWK
 
 | | Crystal structure of pSLA-1*0401 complex with FMDV-derived epitope MTAHITVPY | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
8CSD
 
 | | WbbB D232C Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSE
 
 | | WbbB in complex with alpha-Rha-(1-3)-beta-GlcNAc acceptor | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, N-(8-hydroxyoctyl)-4-methoxybenzamide, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSF
 
 | | WbbB D232C-Kdo adduct + alpha-Rha(1,3)GlcNAc ternary complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSC
 
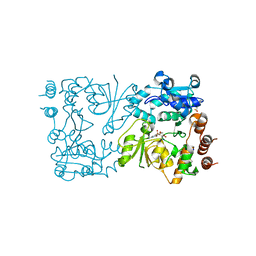 | | WbbB D232N-Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSB
 
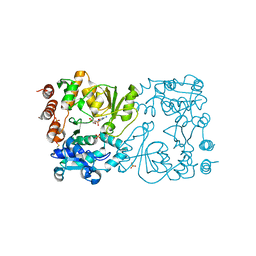 | | WbbB D232N in complex with CMP-beta-Kdo | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
6E16
 
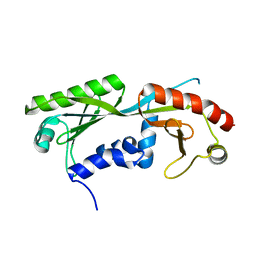 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-09 | | Release date: | 2019-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6E7G
 
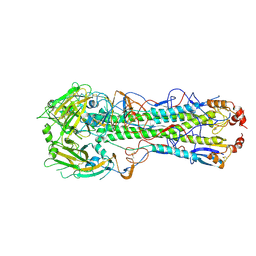 | |
6EKH
 
 | |
7WVP
 
 | |
7WVQ
 
 | |
6E7H
 
 | |
6EJG
 
 | |
5U84
 
 | | Acid ceramidase (ASAH1, aCDase) from common minke whale, Cys143Ala, uncleaved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, ... | | Authors: | Gebai, A, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the activation of acid ceramidase.
Nat Commun, 9, 2018
|
|
5U7Z
 
 | | Human acid ceramidase (ASAH1, aCDase) self-activated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid ceramidase, SULFATE ION, ... | | Authors: | Gebai, A, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the activation of acid ceramidase.
Nat Commun, 9, 2018
|
|
5ULS
 
 | | Structure of GRP94 in the active conformation | | Descriptor: | Endoplasmin, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Huck, J.D, Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.622 Å) | | Cite: | Structural and Functional Analysis of GRP94 in the Closed State Reveals an Essential Role for the Pre-N Domain and a Potential Client-Binding Site.
Cell Rep, 20, 2017
|
|
4HK7
 
 | | Crystal structure of Cordyceps militaris IDCase in complex with uracil | | Descriptor: | URACIL, Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HK5
 
 | | Crystal structure of Cordyceps militaris IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
6JIV
 
 | | SspE crystal structure | | Descriptor: | SspE protein | | Authors: | Bing, Y.Z, Yang, H.G. | | Deposit date: | 2019-02-23 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
4HJW
 
 | | Crystal structure of Metarhizium anisopliae IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
