7W98
 
 | | SARS-CoV-2 Delta S-ACE2-C1 | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9B
 
 | | SARS-CoV-2 Delta S-ACE2-C2b | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W92
 
 | | Open state of SARS-CoV-2 Delta variant spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9F
 
 | | SARS-CoV-2 Delta S-RBD-8D3 | | Descriptor: | Spike protein S1, The heavy chain of 8D3, The light chain of 8D3 | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9I
 
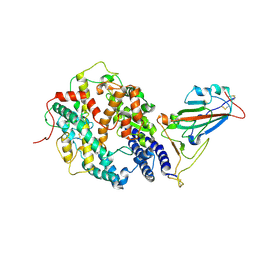 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9E
 
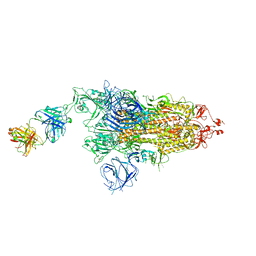 | | SARS-CoV-2 Delta S-8D3 | | Descriptor: | Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain, Spike glycoprotein, The light chain of 8D3 fab | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7WD0
 
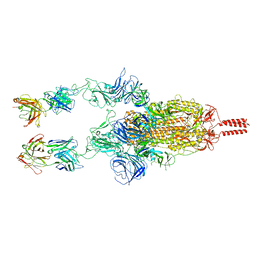 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
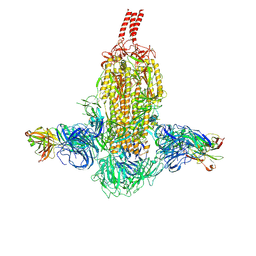 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | | SARS-CoV-2 Beta spike in complex with three S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD7
 
 | | SARS-CoV-2 Beta spike in complex with three S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCZ
 
 | | SARS-CoV-2 Beta spike in complex with one S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
 | | RBD-1 of SARS-CoV-2 Beta spike in complex with S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7DF3
 
 | | SARS-CoV-2 S trimer, S-closed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
6JPE
 
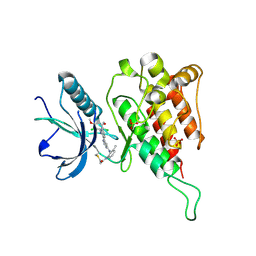 | | Crystal structure of FGFR4 kinase domain with irreversible inhibitor 1 | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[6-[2-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]amino]pyridin-3-yl]pyrimidin-4-yl]amino]-3-methyl-phenyl]prop-2-enamide, SULFATE ION | | Authors: | Chen, X, Dai, S, Zhou, Z, Chen, Y. | | Deposit date: | 2019-03-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Development of a Potent and Specific FGFR4 Inhibitor for the Treatment of Hepatocellular Carcinoma.
J.Med.Chem., 63, 2020
|
|
7YP3
 
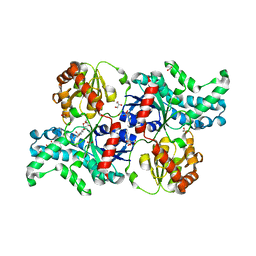 | | Crystal structure of elaiophylin glycosyltransferase in complex with elaiophylin | | Descriptor: | ACETATE ION, Elaiophylin, GLYCEROL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP6
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with UDP | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP5
 
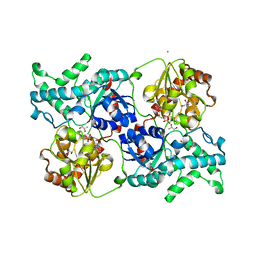 | | Crystal structure of elaiophylin glycosyltransferase in complex with TDP | | Descriptor: | CHLORIDE ION, Glycosyltransferase, R-1,2-PROPANEDIOL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP4
 
 | | Crystal structure of elaiophylin glycosyltransferase in apo-form | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6KFW
 
 | |
8IUO
 
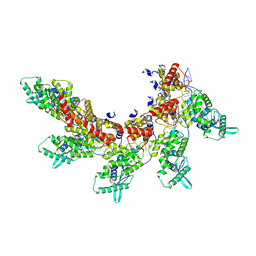 | | respiratory syncytial virus nucleocapsid-like assembly | | Descriptor: | Nucleoprotein, RNA (35-MER) | | Authors: | Wang, Y, Luo, Y, Ling, X, Luo, B, Jia, G, Dong, H, Su, Z. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of the nucleocapsid-like assembly of respiratory syncytial virus.
Signal Transduct Target Ther, 8, 2023
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8JPX
 
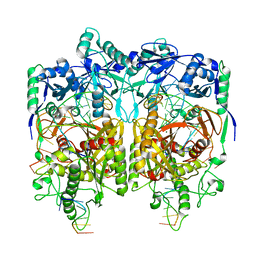 | | Cryo-EM structure of PfAgo-guide DNA-target DNA complex | | Descriptor: | Excess DNA, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
8Y6U
 
 | |
7VCI
 
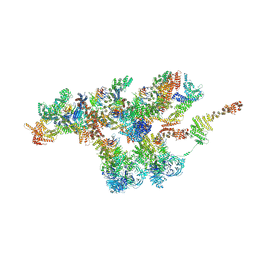 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
8X2F
 
 | | Cryo-EM structure of H5 hemagglutinin from human-infecting H5N8 influenza virus in complex with Sialyl-Lewis X | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Jin, X.Y, Han, P, Song, H, Qi, J.X. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Enhanced binding of Sialyl Lewis X to hemagglutinin contributes to H5N8 mammalian pathogenicity and infectivity
To Be Published
|
|
