7QL8
 
 | | SARS-COV2 Main Protease in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7QG5
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]-6-pyrimidin-5-yl-pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG2
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 6-methyl-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.031 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG3
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 6-[(2~{S})-2-fluoranylpropyl]-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG1
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, methyl 4-[4-[[6-(cyanomethyl)-2-[(1-methylpyrazol-4-yl)amino]-5-oxidanylidene-pyrido[4,3-d]pyrimidin-4-yl]amino]cyclohexyl]piperazine-1-carboxylate | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
5QIL
 
 | |
4Y5T
 
 | | Structure of FtmOx1 apo with metal Iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
5YLX
 
 | | Integrated illustration of a valid epitope based on the SLA class I structure and tetramer technique could carry forward the development of molecular vaccine in swine species | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, PRRSV-NSP9-TMP9 peptide | | Authors: | Pan, X.C, Wei, X.H, Zhang, N, Xia, C. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Illumination of PRRSV Cytotoxic T Lymphocyte Epitopes by the Three-Dimensional Structure and Peptidome of Swine Lymphocyte Antigen Class I (SLA-I).
Front Immunol, 10, 2019
|
|
4Y5S
 
 | | Structure of FtmOx1 with a-Ketoglutarate as co-substrate | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
5QIK
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide | | Descriptor: | GLYCEROL, N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2018-08-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of 4-Azaindole Inhibitors of TGF beta RI as Immuno-oncology Agents.
ACS Med Chem Lett, 9, 2018
|
|
5GTM
 
 | | Modified human MxA, nucleotide-free form | | Descriptor: | Interferon-induced GTP-binding protein Mx1 | | Authors: | Chen, Y, Gao, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Conformational dynamics of dynamin-like MxA revealed by single-molecule FRET
Nat Commun, 8, 2017
|
|
5DDS
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDW
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDU
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
7CN8
 
 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN4
 
 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
6O6L
 
 | | The Structure of EgtB(Cabther) in complex with Hercynine | | Descriptor: | EgtB (Cabther), FE (III) ION, N,N,N-trimethyl-histidine | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
6O6M
 
 | | The Structure of EgtB (Cabther) | | Descriptor: | EgtB (Cabther), FE (III) ION, GLYCEROL | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
5ZXV
 
 | |
5WXB
 
 | |
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | Descriptor: | Dipeptide permease D | | Authors: | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
8CYF
 
 | | WhiB3 bound to SigmaAr4-RNAP Beta flap tip chimera and DNA | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*CP*AP*CP*AP*AP*CP*CP*GP*AP*TP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*TP*CP*GP*GP*TP*TP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Wan, T, Zhang, L.M. | | Deposit date: | 2022-05-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis of DNA binding by the WhiB-like transcription factor WhiB3 in Mycobacterium tuberculosis.
J.Biol.Chem., 299, 2023
|
|
4YWC
 
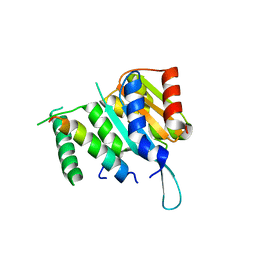 | | Crystal structure of Myc3(5-242) fragment in complex with Jaz9(218-239) peptide | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2015-03-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4YVV
 
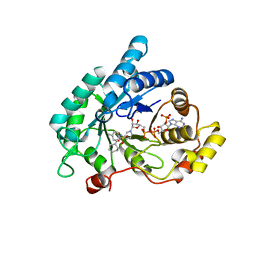 | | Crystal structure of AKR1C3 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
6VPX
 
 | |
