6ED1
 
 | | Bacteroides dorei Beta-glucuronidase | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein, SODIUM ION | | Authors: | Biernat, K.A, Redinbo, M.R. | | Deposit date: | 2018-08-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
6EC6
 
 | | Ruminococcus gnavus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Redinbo, M.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
3P0I
 
 | |
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q4S
 
 | |
3P0H
 
 | | Leishmania major Tyrosyl-tRNA synthetase in complex with fisetin, cubic crystal form | | Descriptor: | 3,7,3',4'-TETRAHYDROXYFLAVONE, Tyrosyl-tRNA synthetase | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Double-Length Tyrosyl-tRNA Synthetase from the Eukaryote Leishmania major Forms an Intrinsically Asymmetric Pseudo-Dimer.
J.Mol.Biol., 409, 2011
|
|
8U02
 
 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-28 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
8TZQ
 
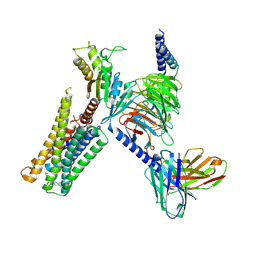 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE Mutant, scFv16, and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-27 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
8UMG
 
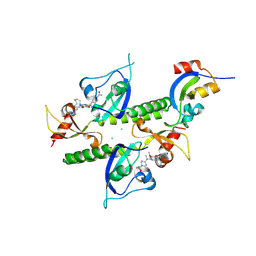 | | Chromodomains of human CHD1 complexed with UNC10142 | | Descriptor: | 1-{4-[{2-(azonan-1-yl)-6-methoxy-7-[3-(piperidin-1-yl)propoxy]quinazolin-4-yl}(methyl)amino]piperidin-1-yl}ethan-1-one, CHLORIDE ION, Chromodomain-helicase-DNA-binding protein 1 | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Optimization of a quinazoline-based scaffold against human CHD1
To Be Published
|
|
8QOB
 
 | |
3P0J
 
 | |
4Z6G
 
 | | Structure of NT domain | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, PHOSPHATE ION | | Authors: | Yang, F, Zhang, Y. | | Deposit date: | 2015-04-05 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun, 7, 2016
|
|
3S84
 
 | | Dimeric apoA-IV | | Descriptor: | Apolipoprotein A-IV, SULFATE ION | | Authors: | Deng, X, Davidson, W.S, Thompson, T.B. | | Deposit date: | 2011-05-27 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Dimeric Apolipoprotein A-IV and Its Mechanism of Self-Association.
Structure, 20, 2012
|
|
3D4G
 
 | |
5C25
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((4-((4-cyanophenyl)amino)-1,3,5-triazin-2-yl)amino)-5,7-dimethyl-2-naphthonitrile (JLJ639), a Non-nucleoside Inhibitor | | Descriptor: | 6-({4-[(4-cyanophenyl)amino]-1,3,5-triazin-2-yl}amino)-5,7- dimethyl-2-naphthonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Frey, K.M, Anderson, K.S. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery and crystallography of bicyclic arylaminoazines as potent inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3SX9
 
 | |
5C24
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-((4-((4-cyanophenyl)amino)-1,3,5-triazin-2-yl)amino)-6,8-dimethylindolizine-2-carbonitrile (JLJ605), a non-nucleoside inhibitor | | Descriptor: | 6-({4-[(4-cyanophenyl)amino]-1,3,5-triazin-2-yl}amino)-5,7-dimethylindolizine-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and crystallography of bicyclic arylaminoazines as potent inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOS
 
 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5C42
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K101P) Variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, HIV-1 Reverse Transcriptase, p51 subunit, ... | | Authors: | Frey, K.M, Gray, W.T, Anderson, K.S. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Inhibitors Active against HIV Reverse Transcriptase with K101P, a Mutation Conferring Rilpivirine Resistance.
Acs Med.Chem.Lett., 6, 2015
|
|
3STB
 
 | | A complex of two editosome proteins and two nanobodies | | Descriptor: | MP18 RNA editing complex protein, RNA-editing complex protein MP42, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody.
Nucleic Acids Res., 40, 2012
|
|
3TN2
 
 | | structure analysis of MIP1-beta P8A | | Descriptor: | C-C motif chemokine 4, ZINC ION | | Authors: | Guo, Q, Tang, W.J. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
3I7C
 
 | |
3IBE
 
 | | Crystal Structure of a Pyrazolopyrimidine Inhibitor Bound to PI3 Kinase Gamma | | Descriptor: | 1-(4-{4-morpholin-4-yl-1-[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}phenyl)-3-pyridin-4-ylurea, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | ATP-Competitive Inhibitors of the Mammalian Target of Rapamycin: Design and Synthesis of Highly Potent and Selective Pyrazolopyrimidines.
J.Med.Chem., 52, 2009
|
|
3I7F
 
 | |
