2KSZ
 
 | |
5XFA
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
8GNN
 
 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
6IGX
 
 | | Crystal structure of human CAP-G in complex with CAP-H | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Condensin complex subunit 2, Condensin complex subunit 3 | | Authors: | Hara, K, Migita, T, Shimizu, K, Hashimoto, H. | | Deposit date: | 2018-09-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural basis of HEAT-kleisin interactions in the human condensin I subcomplex.
Embo Rep., 20, 2019
|
|
5H4J
 
 | | Crystal structure of Human dUTPase in complex with N-[(1R)-1-[3-(Cyclopentyloxy)-phenyl]-ethyl]-3-[(3,4-dihydro-2,4-dioxo-1(2H)-pyrimidinyl)methoxy]-1-propanesulfonamide | | Descriptor: | DIMETHYL SULFOXIDE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, ... | | Authors: | Chong, K.T, Miyahara, S, Miyakoshi, H, Fukuoka, M. | | Deposit date: | 2016-11-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TAS-114, a First-in-Class Dual dUTPase/DPD Inhibitor, Demonstrates Potential to Improve Therapeutic Efficacy of Fluoropyrimidine-Based Chemotherapy.
Mol. Cancer Ther., 17, 2018
|
|
6IRZ
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
5XF9
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
6IRV
 
 | |
6IS0
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRW
 
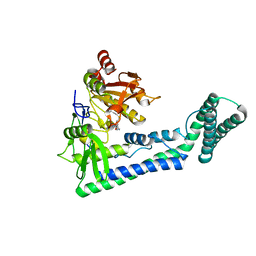 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
7TIA
 
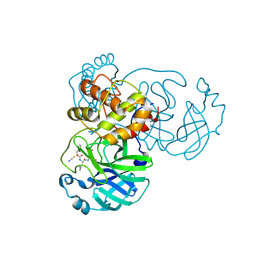 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-14 | | Descriptor: | 3C-like proteinase nsp5, THIOCYANATE ION, benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TJ0
 
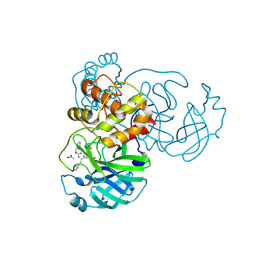 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
6IRY
 
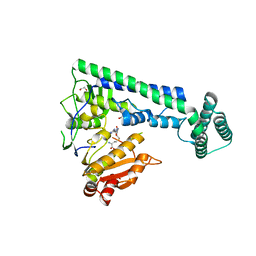 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRX
 
 | |
2RQ2
 
 | | The solution structure of the N-terminal fragment of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Mizuguchi, M, Aizawa, T, Shinoda, H, Demura, M, Kawabata, S, Kawano, K. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: big defensin changes its N-terminal structure to associate with the target membrane
Biochemistry, 48, 2009
|
|
7V5N
 
 | | Crystal structure of Fab fragment of bevacizumab bound to DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*GP*TP*TP*GP*GP*TP*GP*GP*TP*AP*GP*TP*TP*AP*CP*GP*TP*TP*CP*GP*C)-3'), IMIDAZOLE, ... | | Authors: | Hishiki, A, Tong, J, Todoroki, K, Hashimoto, H. | | Deposit date: | 2021-08-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Biosens.Bioelectron., 203, 2022
|
|
6IQM
 
 | | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase Complexed with NAD+ from Lactobacillus plantarum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Yoneda, K, Kinoshita, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Milk Sci, 68, 2019
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6IQV
 
 | | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase Complexed with Hg2+ from Lactobacillus plantarum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Yoneda, K, Kinoshita, H. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Milk Sci, 68, 2019
|
|
2X1C
 
 | | The crystal structure of precursor acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | ACYL-COENZYME, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
7FI3
 
 | | Archaeal oligopeptide permease A (OppA) from Thermococcus kodakaraensis in complex with an endogenous pentapeptide | | Descriptor: | ABC-type dipeptide/oligopeptide transport system, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Yokoyama, H, Kamei, N, Konishi, K, Hara, K, Hashimoto, H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptide recognition by archaeal oligopeptide permease A.
Proteins, 90, 2022
|
|
2X1D
 
 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, ACETATE ION, ACYL-COENZYME, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
6MK9
 
 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121
To Be Published
|
|
6MKL
 
 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function analysis of darunavir-resistant-P51 HIV-1 protease in complex with GRL-142.
To Be Published
|
|
