7BZA
 
 | | Template lasso peptide C24 | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
2NAX
 
 | | Structure of CCHC zinc finger domain of Pcf11 | | Descriptor: | Protein PCF11, ZINC ION | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C terminus of Pcf11 forms a novel zinc-finger structure that plays an essential role in mRNA 3'-end processing.
RNA, 23, 2017
|
|
3QC4
 
 | | PDK1 in complex with DFG-OUT inhibitor xxx | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Arndt, J.W. | | Deposit date: | 2011-01-15 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8FAR
 
 | |
7T3V
 
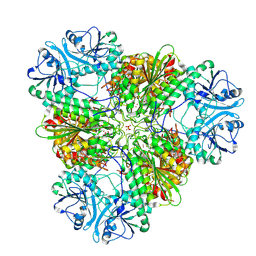 | | Metal dependent activation of Plasmodium falciparum M17 aminopeptidase, spacegroup P22121 after crystals soaked with Zn2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
J.Biol.Chem., 298, 2022
|
|
7SRV
 
 | | Metal dependent activation of Plasmodium falciparum M17 aminopeptidase (inactive form), spacegroup P22121 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
J.Biol.Chem., 298, 2022
|
|
7TO4
 
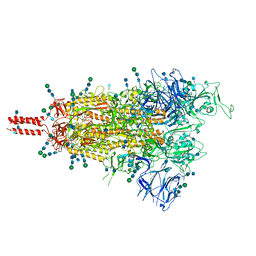 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
7TNW
 
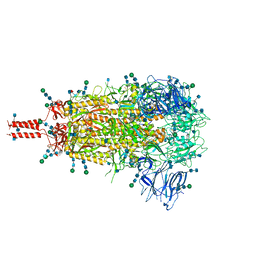 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-21 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
5UZZ
 
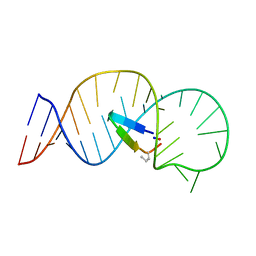 | |
5WAL
 
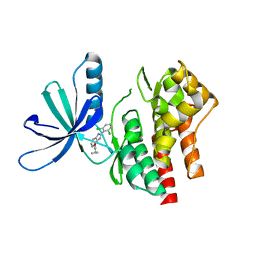 | |
5UZT
 
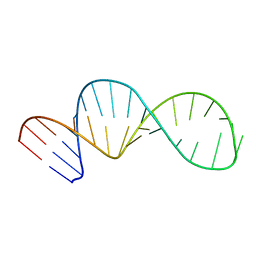 | |
5WH6
 
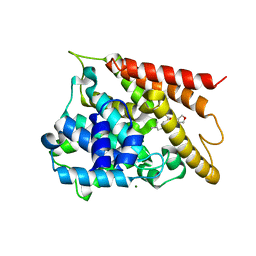 | | Crystal structure of PDE4D2 in complex with inhibitor (S_Zl-n-91) | | Descriptor: | 1-[4-(difluoromethoxy)-3-{[(3S)-oxolan-3-yl]oxy}phenyl]-3-methylbutan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H, Wang, H. | | Deposit date: | 2017-07-14 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a PDE4-Specific Pocket for the Design of Selective Inhibitors.
Biochemistry, 57, 2018
|
|
5WEV
 
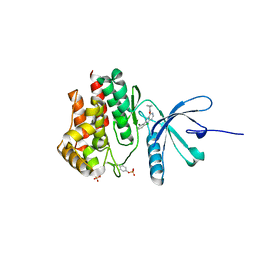 | |
6AUD
 
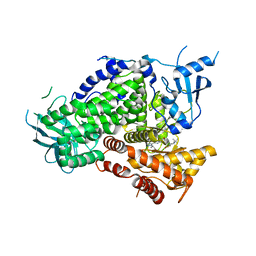 | | PI3K-gamma K802T in complex with Cpd 8 10-((1-(tert-butyl)piperidin-4-yl)sulfinyl)-2-(1-isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepine | | Descriptor: | 10-[(S)-(1-tert-butylpiperidin-4-yl)sulfinyl]-2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Ultsch, M. | | Deposit date: | 2017-08-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Design of Selective Benzoxazepin PI3K delta Inhibitors Through Control of Dihedral Angles.
ACS Med Chem Lett, 8, 2017
|
|
3TYQ
 
 | |
8F6R
 
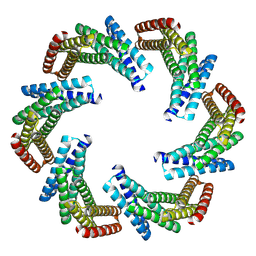 | | CryoEM structure of designed modular protein oligomer C6-79 | | Descriptor: | De novo designed oligomeric protein C6-79 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
4TLG
 
 | | Crystal structure of SEC14-like protein 4 (SEC14L4) | | Descriptor: | 1,2-ETHANEDIOL, SEC14-like protein 4, UNDECANOIC ACID | | Authors: | Vollmar, M, Kopec, J, Kiyani, W, Shrestha, L, Sorrell, F, Krojer, T, Williams, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of SEC14-like protein 4 (SEC14L4)
To Be Published
|
|
7CNN
 
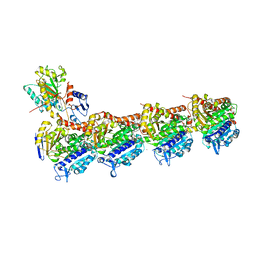 | | vinorelbine in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7CNO
 
 | | Phomopsin A in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7CNM
 
 | | YDX in complex with tubulin | | Descriptor: | (2~{S},4~{S})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-4-methyl-3-[[(2~{S},3~{S})-3-methyl-2-[[(2~{R})-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-cyclohexyl-2-methyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
8J00
 
 | | Human KCNQ2-CaM in complex with CBD | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, cannabidiol | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8IZY
 
 | | Human KCNQ2-CaM in complex with HN37 | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2, methyl N-[4-[(4-fluorophenyl)methyl-prop-2-ynyl-amino]-2,6-dimethyl-phenyl]carbamate | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8JM9
 
 | |
