7CZZ
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2F11_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D00
 
 | | S protein of SARS-CoV-2 in complex bound with FabP5A-1B8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,chain H of FabP5A-1B8,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZW
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2G7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D03
 
 | | S protein of SARS-CoV-2 in complex bound with FabP5A-2G7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IGL c2312_light_IGLV2-14_IGLJ2,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0D
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | Authors: | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
6E4R
 
 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9B | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
6E4Q
 
 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9A in Complex with UDP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
6A2B
 
 | | Crystal Structure of Xenopus laevis MHC I complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, TYR-MET-MET-PRO-ARG-HIS-TRP-PRO-ILE | | Authors: | Ma, L.Z, Xia, C. | | Deposit date: | 2018-06-10 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Glimpse of the Peptide Profile Presentation byXenopus laevisMHC Class I: Crystal Structure of pXela-UAA Reveals a Distinct Peptide-Binding Groove.
J Immunol., 204, 2020
|
|
6AJG
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SQ109 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, DODECYL-BETA-D-MALTOSIDE, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJJ
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with ICA38 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4,6-difluoro-N-(spiro[5.5]undecan-3-yl)-1H-indole-2-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
3RDH
 
 | | X-ray induced covalent inhibition of 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | Authors: | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | Deposit date: | 2011-04-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8IW9
 
 | | Cryo-EM structure of the CAD-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWM
 
 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | Descriptor: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWE
 
 | | Cryo-EM structure of the SPE-mTAAR9 complex | | Descriptor: | SPERMIDINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
4M84
 
 | |
1QP9
 
 | | STRUCTURE OF HAP1-PC7 COMPLEXED TO THE UAS OF CYC7 | | Descriptor: | CYP1(HAP1-PC7) ACTIVATORY PROTEIN, DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), ... | | Authors: | Lukens, A, King, D, Marmorstein, R. | | Deposit date: | 1999-06-01 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.
Nucleic Acids Res., 28, 2000
|
|
8JXZ
 
 | | Chitin binding SusD-like protein AqSusD in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SusD-like protein AqSusD | | Authors: | Yang, J. | | Deposit date: | 2023-07-01 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
4DHF
 
 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5YLX
 
 | | Integrated illustration of a valid epitope based on the SLA class I structure and tetramer technique could carry forward the development of molecular vaccine in swine species | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, PRRSV-NSP9-TMP9 peptide | | Authors: | Pan, X.C, Wei, X.H, Zhang, N, Xia, C. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Illumination of PRRSV Cytotoxic T Lymphocyte Epitopes by the Three-Dimensional Structure and Peptidome of Swine Lymphocyte Antigen Class I (SLA-I).
Front Immunol, 10, 2019
|
|
8GZ8
 
 | | Cryo-EM structure of Abeta2 fibril polymorph1 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
8GZ9
 
 | | Cryo-EM structure of Abeta2 fibril polymorph2 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
5DDW
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
8H77
 
 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
5ZXV
 
 | |
7YRD
 
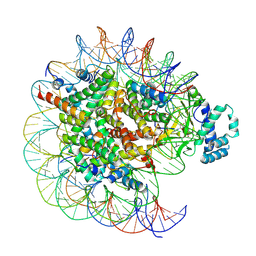 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
