4Z82
 
 | | Cysteine bound rat cysteine dioxygenase C164S variant at pH 8.1 | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Siakkou, E, Rutledge, M.T, Kanitz, M, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2015-04-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
J.Biol.Inorg.Chem., 21, 2016
|
|
4ZPV
 
 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
4ZPW
 
 | | Structure of unbound MERS-CoV spike receptor-binding domain (England1 strain). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Spike glycoprotein | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
1CYD
 
 | | CARBONYL REDUCTASE COMPLEXED WITH NADPH AND 2-PROPANOL | | Descriptor: | CARBONYL REDUCTASE, ISOPROPYL ALCOHOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family.
Structure, 4, 1996
|
|
5A7C
 
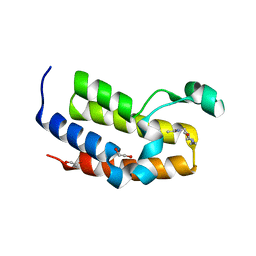 | | Crystal structure of the second bromodomain of human BRD3 in complex with compound | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 3, N-(6-ACETAMIDOHEXYL)ACETAMIDE | | Authors: | Welin, M, Kimbung, R, Diehl, C, Hakansson, M, Logan, D.T, Walse, B. | | Deposit date: | 2015-07-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cancer Differentiating Agent Hexamethylene Bisacetamide Inhibits Bet Bromodomain Proteins.
Cancer Res., 76, 2016
|
|
4C66
 
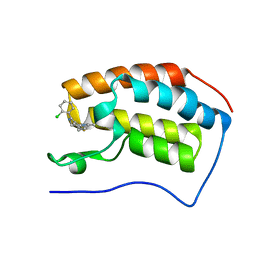 | | Discovery of Epigenetic Regulator I-BET762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the BET Bromodomains | | Descriptor: | 4-(2-chlorophenyl)-2-ethyl-9-methyl-6,8-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Epigenetic Regulator I-Bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains.
J.Med.Chem., 56, 2013
|
|
4C67
 
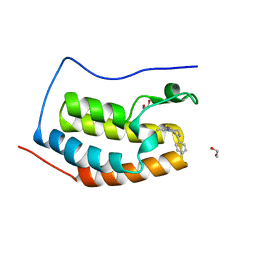 | | Discovery of Epigenetic Regulator I-BET762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the BET Bromodomains | | Descriptor: | 1,2-ETHANEDIOL, 13-methyl-7-phenyl-3-thia-1,8,11,12-tetraazatricyclo trideca-2(6),4,7,10,12-pentaene, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Epigenetic Regulator I-Bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains.
J.Med.Chem., 56, 2013
|
|
5KQE
 
 | |
1BJN
 
 | |
8TP6
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 4-1-1E02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 4-1-1E02 Fab, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP7
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with Sa-targeting Fab 4-1-1G03 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 4-1-1G03, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP9
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TPA
 
 | | H1 hemagglutinin (NC99) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP3
 
 | | H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1F05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1F05, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP4
 
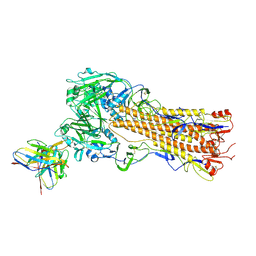 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 1-1-1E04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1E04, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP2
 
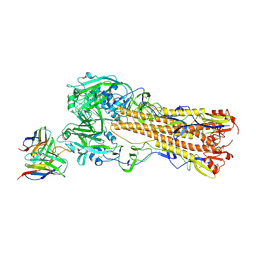 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting 1-1-1F05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1F05, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP5
 
 | | H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1E04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1E04, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
5Y2X
 
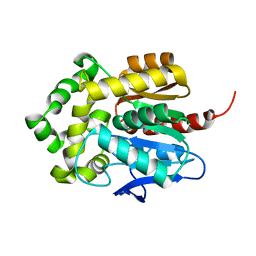 | | Crystal structure of apo-HaloTag (M175C) | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
5YLO
 
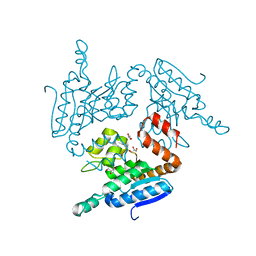 | | Structural of Pseudomonas aeruginosa PA4980 | | Descriptor: | GLYCEROL, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Li, T, Peng, C.T, Li, C.C, Xiao, Q.J, He, L.H, Wang, N.Y, Bao, R. | | Deposit date: | 2017-10-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural characterization of a Delta3, Delta2-enoyl-CoA isomerase from Pseudomonas aeruginosa: implications for its involvement in unsaturated fatty acid metabolism.
J.Biomol.Struct.Dyn., 37, 2019
|
|
2D5I
 
 | |
8I5I
 
 | |
8I5H
 
 | |
5Y2Y
 
 | | Crystal structure of HaloTag (M175C) complexed with dansyl-PEG2-HaloTag ligand | | Descriptor: | 5-(dimethylamino)-~{N}-[2-(2-hexoxyethoxy)ethyl]naphthalene-1-sulfonamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
6QX4
 
 | |
1KEG
 
 | | Antibody 64M-2 Fab complexed with dTT(6-4)TT | | Descriptor: | 5'-D(*TP*(64T)P*TP*T)-3', Anti-(6-4) photoproduct antibody 64M-2 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-2 Fab (light chain), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Sato, K, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2001-11-15 | | Release date: | 2002-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DNA (6-4) photoproduct dTT(6-4)TT in complex with the 64M-2 antibody Fab fragment implies increased antibody-binding affinity by the flanking nucleotides.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
