7WFC
 
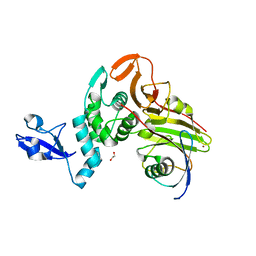 | | X-ray structure of HKU1-PLP2(Cys109Ser) catalytic mutant in complex with free ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, 60S ribosomal protein L40, Papain-like protease, ... | | Authors: | Xiong, Y.X, Fu, Z.Y, Huang, H. | | Deposit date: | 2021-12-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression.
Sci.Signal., 16, 2023
|
|
3CMU
 
 | |
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
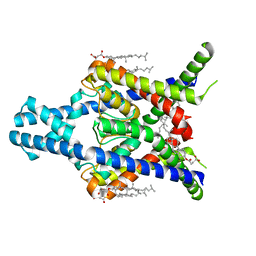
8K1Q
 
 | |
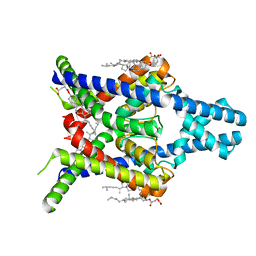
8K1V
 
 | |
8HJ5
 
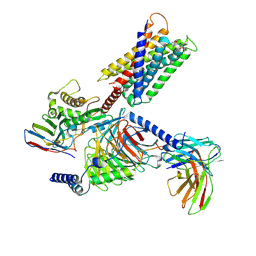 | | Cryo-EM structure of Gq-coupled MRGPRX1 bound with Compound-16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Gan, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of agonist-induced activation of the human itch receptor MRGPRX1.
Plos Biol., 21, 2023
|
|
8K1J
 
 | |
8K1Z
 
 | |
4K3X
 
 | |
3QKD
 
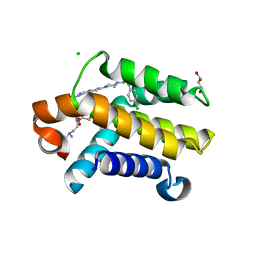 | | Crystal structure of Bcl-xL in complex with a Quinazoline sulfonamide inhibitor | | Descriptor: | (R)-N-(7-(4-((4'-chlorobiphenyl-2-yl)methyl)piperazin-1-yl)quinazolin-4-yl)-4-(4-(dimethylamino)-1-(phenylthio)butan-2-ylamino)-3-nitrobenzenesulfonamide, Bcl-2-like protein 1, CHLORIDE ION, ... | | Authors: | Czabotar, P.E, Smith, B.J. | | Deposit date: | 2011-01-31 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity.
J.Med.Chem., 54, 2011
|
|
7V7M
 
 | | crystal structure of SARS-CoV-2 3CL protease | | Descriptor: | 3C-like proteinase | | Authors: | Yi, Y, Zhang, M, Ye, M. | | Deposit date: | 2021-08-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4JA8
 
 | | Complex of Mitochondrial Isocitrate Dehydrogenase R140Q Mutant with AGI-6780 Inhibitor | | Descriptor: | 1-[5-(cyclopropylsulfamoyl)-2-thiophen-3-yl-phenyl]-3-[3-(trifluoromethyl)phenyl]urea, CALCIUM ION, GLYCEROL, ... | | Authors: | Wei, W, Chen, L, Wu, M, Jiang, F, Travins, J, Qian, K, DeLaBarre, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation.
Science, 340, 2013
|
|
4K3Y
 
 | |
4O1I
 
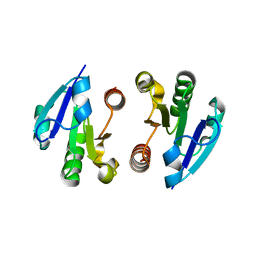 | | Crystal Structure of the regulatory domain of MtbGlnR | | Descriptor: | Transcriptional regulatory protein | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4O1H
 
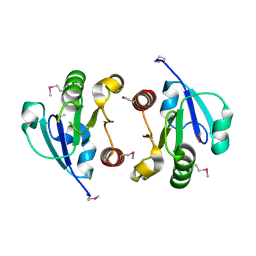 | | Crystal Structure of the regulatory domain of AmeGlnR | | Descriptor: | Transcription regulator GlnR | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4NXL
 
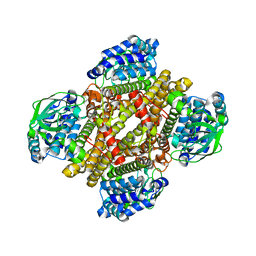 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | DszC | | Authors: | Zhang, L, Duan, X, Li, X, Rao, Z. | | Deposit date: | 2013-12-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
8FZC
 
 | |
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
8GU4
 
 | | Poly(ethylene terephthalate) hydrolase (IsPETase)-linker | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
