1PPC
 
 | | GEOMETRY OF BINDING OF THE BENZAMIDINE-AND ARGININE-BASED INHIBITORS N-ALPHA-(2-NAPHTHYL-SULPHONYL-GLYCYL)-DL-P-AMIDINOPHENYLALANYL-PIPERIDINE (NAPAP) AND (2R,4R)-4-METHYL-1-[N-ALPHA-(3-METHYL-1,2,3,4-TETRAHYDRO-8-QUINOLINESULPHONYL)-L-ARGINYL]-2-PIPERIDINE CARBOXYLIC ACID (MQPA) TO HUMAN ALPHA-THROMBIN: X-RAY CRYSTALLOGRAPHIC DETERMINATION OF THE NAPAP-TRYPSIN COMPLEX AND MODELING OF NAPAP-THROMBIN AND MQPA-THROMBIN | | Descriptor: | 1-[N-(naphthalen-2-ylsulfonyl)glycyl-4-carbamimidoyl-D-phenylalanyl]piperidine, CALCIUM ION, TRYPSIN | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Geometry of binding of the benzamidine- and arginine-based inhibitors N alpha-(2-naphthyl-sulphonyl-glycyl)-DL-p-amidinophenylalanyl-pipe ridine (NAPAP) and (2R,4R)-4-methyl-1-[N alpha-(3-methyl-1,2,3,4-tetrahydr quinolinesulphonyl)-L-arginyl]-2-piperidine carboxylic acid (MQPA) to human alpha-thrombin.X-ray crystallographic determination of the NAPAP-trypsin complex and modeling of NAPAP-thrombin and MQPA-thrombin.
Eur.J.Biochem., 193, 1990
|
|
1EF7
 
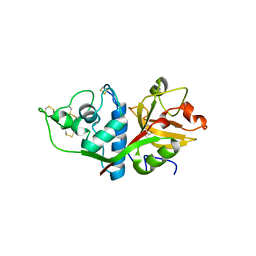 | | CRYSTAL STRUCTURE OF HUMAN CATHEPSIN X | | Descriptor: | CATHEPSIN X | | Authors: | Guncar, G, Klemencic, I, Turk, B, Turk, V, Karaoglanovic-Carmona, A, Juliano, L, Turk, D. | | Deposit date: | 2000-02-07 | | Release date: | 2000-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease.
Structure Fold.Des., 8, 2000
|
|
8PCH
 
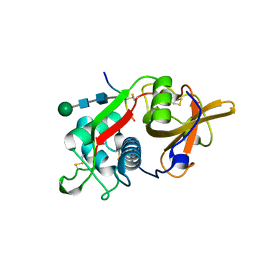 | | CRYSTAL STRUCTURE OF PORCINE CATHEPSIN H DETERMINED AT 2.1 ANGSTROM RESOLUTION: LOCATION OF THE MINI-CHAIN C-TERMINAL CARBOXYL GROUP DEFINES CATHEPSIN H AMINOPEPTIDASE FUNCTION | | Descriptor: | CATHEPSIN H, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guncar, G, Podobnik, M, Pungercar, J, Strukelj, B, Turk, V, Turk, D. | | Deposit date: | 1997-11-07 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of porcine cathepsin H determined at 2.1 A resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function.
Structure, 6, 1998
|
|
7QKD
 
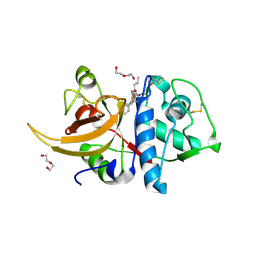 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
8PRX
 
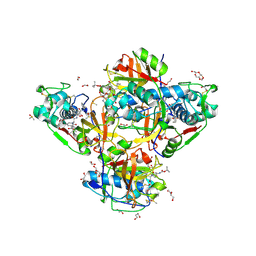 | | Crystal structure of human cathepsin L after reaction with the bound ketoamide inhibitor 13b | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-07-12 | | Release date: | 2023-08-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8OZA
 
 | | Human cathepsin L in complex with covalently bound CA-074 methyl ester | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Ewert, W, Reinke, P.Y.A, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-05-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8OFA
 
 | | Crystal structure of human cathepsin L interacting with tosyl phenylalanyl chloromethyl ketone (TPCK) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 4-methyl-~{N}-[(2~{S})-4-oxidanyl-3-oxidanylidene-1-phenyl-butan-2-yl]benzenesulfonamide, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Ewert, W, Reinke, P.Y.A, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-03-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QKB
 
 | | Crystal structure of human cathepsin L in complex with the vinyl sulfone inhibitor K777 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
7ZS7
 
 | | Crystal structure of human cathepsin L with covalently bound calpain inhibitor VI | | Descriptor: | (2S)-2-[(4-fluorophenyl)sulfonylamino]-3-methyl-N-[(2S)-4-methyl-1-oxidanyl-pentan-2-yl]butanamide, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
7ZXA
 
 | | Crystal structure of human cathepsin L with covalently bound aloxistatin (E-64D) | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4V
 
 | | Crystal structure of human cathepsin L with covalently bound E-64 | | Descriptor: | Cathepsin L, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4W
 
 | | Crystal structure of human cathepsin L with covalently bound Cathepsin L inhibitor IV | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4X
 
 | | Crystal structure of human Cathepsin L with covalently bound Calpain inhibitor III | | Descriptor: | (phenylmethyl) N-[(2S)-3-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]carbamate, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4U
 
 | | Crystal structure of human cathepsin L with CAA0225 | | Descriptor: | (2S,3S)-N3-[2-(4-hydroxyphenyl)ethyl]-N2-[(2S)-1-oxidanylidene-3-phenyl-1-[(phenylmethyl)amino]propan-2-yl]oxirane-2,3-dicarboxamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A5B
 
 | | Crystal structure of human cathepsin L in complex with covalently bound MG-101 | | Descriptor: | Calpain Inhibitor I, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
7ZVF
 
 | | Crystal structure of human cathepsin L in complex with covalently bound CLIK148 | | Descriptor: | (2S)-N-[(2S)-1-(dimethylamino)-1-oxidanylidene-3-phenyl-propan-2-yl]-2-oxidanyl-N'-(2-pyridin-2-ylethyl)butanediamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Tsuge, H, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8C3D
 
 | | Sulfonated Calpeptin is a promising drug candidate against SARS-CoV-2 infections | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CALCIUM ION, Cathepsin K | | Authors: | Loboda, J, Karnicar, K, Lindic, N, Usenik, A, Lieske, J, Meents, A, Guenther, S, Reinke, P.Y.A, Falke, S, Ewert, W, Turk, D. | | Deposit date: | 2022-12-23 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7APH
 
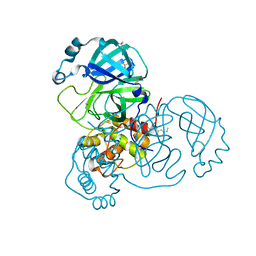 | | Structure of SARS-CoV-2 Main Protease bound to Tofogliflozin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AK4
 
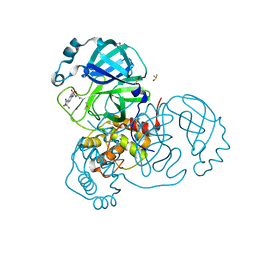 | | Structure of SARS-CoV-2 Main Protease bound to Tretazicar. | | Descriptor: | 3C-like proteinase, 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AGA
 
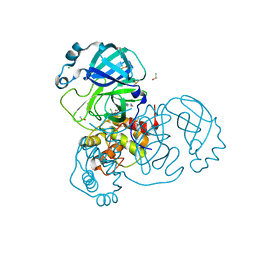 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ANS
 
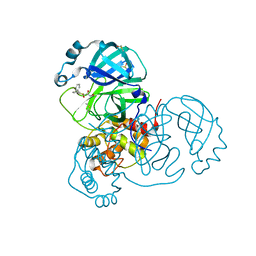 | | Structure of SARS-CoV-2 Main Protease bound to Adrafinil. | | Descriptor: | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
