8G0W
 
 | |
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
5TRV
 
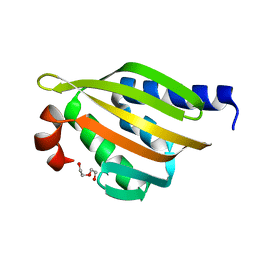 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
4QX5
 
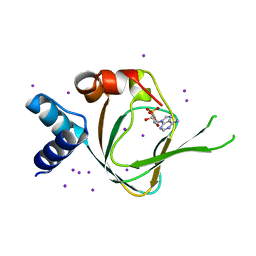 | | Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: Insights for PKG agonist design | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, IODIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Huang, G.Y, Gerlits, O.O, Blakeley, M.P, Sankaran, B, Kovalevsky, A.Y, Kim, C. | | Deposit date: | 2014-07-18 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Neutron Diffraction Reveals Hydrogen Bonds Critical for cGMP-Selective Activation: Insights for cGMP-Dependent Protein Kinase Agonist Design.
Biochemistry, 53, 2014
|
|
4RVA
 
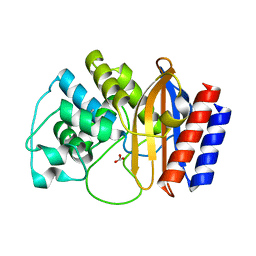 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for deacylation | | Descriptor: | BICARBONATE ION, Beta-lactamase TEM | | Authors: | Stojanoski, V, Chow, D.-C, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4397 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX3
 
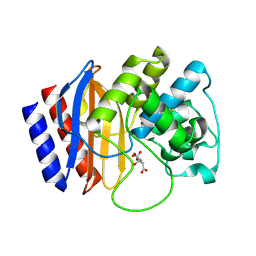 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | Descriptor: | Beta-lactamase TEM, CITRATE ANION | | Authors: | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX2
 
 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | Descriptor: | Beta-lactamase TEM, SULFATE ION | | Authors: | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4W9N
 
 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase complexed with triclosan | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, IMIDAZOLE, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
5BW2
 
 | | X-ray crystal structure of Aplysia californica acetylcholine binding protein (Ac-AChBP) Y55W in complex with 2-Pyridin-3-yl-1-aza-bicyclo[2.2.2]octane; 2-(3-pyridyl)quinuclidine; 2-PQ (TI-4699) | | Descriptor: | (2R)-2-(pyridin-3-yl)-1-azabicyclo[2.2.2]octane, Soluble acetylcholine receptor | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-06-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
5C6C
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cAMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CADMIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-22 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5C67
 
 | |
5C6X
 
 | | Crystal structure of C-As lyase with Co(II) | | Descriptor: | COBALT (II) ION, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of C-As lyase with Co(II)
To Be Published
|
|
5CB9
 
 | | Crystal structure of C-As lyase with mercaptoethonal | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
5C8W
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MALONIC ACID, SODIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5DRH
 
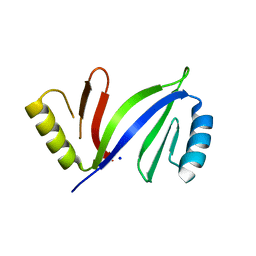 | | Crystal structure of apo C-As lyase | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, SODIUM ION | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of apo C-As lyase
To Be Published
|
|
5DFG
 
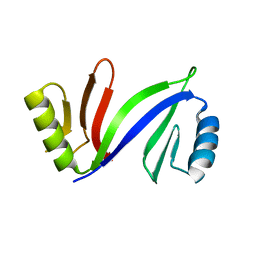 | |
5D4F
 
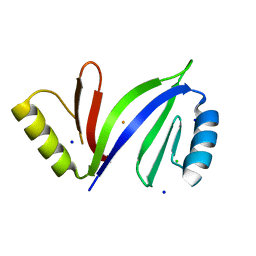 | | Crystal structure of C-As lyase with Fe(III) | | Descriptor: | CHLORIDE ION, FE (III) ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-08-07 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
8TMT
 
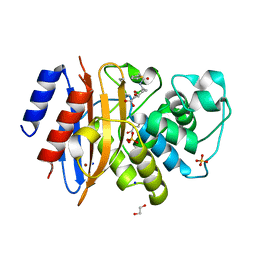 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJM
 
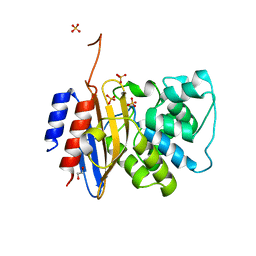 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TMR
 
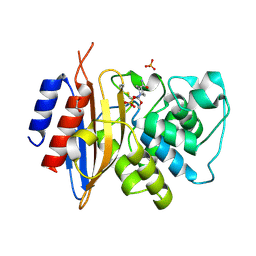 | | Crystal structure of KPC-44 carbapenemase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TN0
 
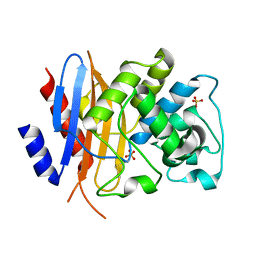 | | Crystal structure of KPC-44 carbapenemase w/o cryoprotectant | | Descriptor: | SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
5EVJ
 
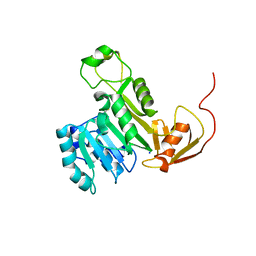 | | X-ray crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii | | Descriptor: | Arsenite methyltransferase, SODIUM ION | | Authors: | Packianathan, C, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii
To Be Published
|
|
5L33
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
4PMA
 
 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase at 1.39 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Cardenas, A.M, Adamski, C.J, Brown, N.G, Horton, L.B, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
