6AE4
 
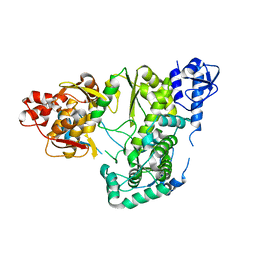 | |
4MSV
 
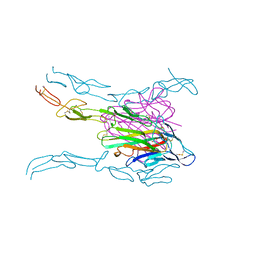 | | Crystal structure of FASL and DcR3 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 6, ... | | Authors: | Liu, W, Ramagopal, U.A, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
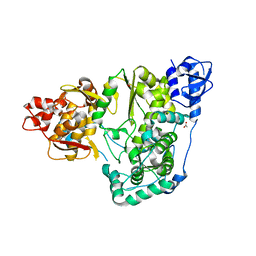
6AE6
 
 | |
6AE7
 
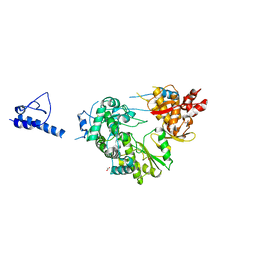 | |
6AE5
 
 | |
4KG8
 
 | | Crystal structure of light mutant | | Descriptor: | Tumor necrosis factor ligand superfamily member 14 | | Authors: | Liu, W, Zhan, C, Kumar, P.R, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-28 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-29 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4NC3
 
 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
4KGG
 
 | | Crystal structure of light mutant2 and dcr3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Bonanno, J.B, Zhan, C, Kumar, P.R, Toro, R, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
8HZQ
 
 | | Bacillus subtilis SepF protein assembly (wild type) | | Descriptor: | Cell division protein SepF | | Authors: | Liu, W. | | Deposit date: | 2023-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular basis for curvature formation in SepF polymerization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8HZT
 
 | |
6NG3
 
 | | Crystal structure of human CD160 and HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | Authors: | Liu, W, Bonanno, J, Almo, S.C. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
6NG9
 
 | |
5YF8
 
 | |
5YF6
 
 | |
5YF7
 
 | |
5YF5
 
 | |
8WOR
 
 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOT
 
 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOQ
 
 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOS
 
 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
3D6V
 
 | | Crystal structure of 4-(trifluoromethyldiazirinyl)phenylalanyl-tRNA synthetase | | Descriptor: | 4-(2,2,2-TRIFLUOROETHYL)-L-PHENYLALANINE, BETA-MERCAPTOETHANOL, Tyrosyl-tRNA synthetase | | Authors: | Liu, W, Tippmann, E, Mack, A.V, Schultz, P.G. | | Deposit date: | 2008-05-20 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A genetically encoded diazirine photocrosslinker in Escherichia coli
ChemBioChem, 8, 2007
|
|
7BZ0
 
 | | complex structure of alginate lyase AlyF-OU02 with G6 | | Descriptor: | Alginate lyase AlyF-OU02, CALCIUM ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Liu, W, Lyu, Q, Zhang, K. | | Deposit date: | 2020-04-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate-binding cleft of AlyF reveal the first long-chain alginate-binding mode.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8I7L
 
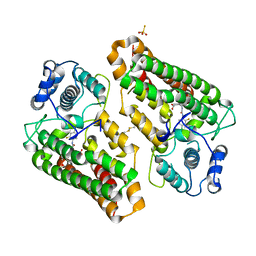 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with a novel inhibitor | | Descriptor: | 1-[3-[(4-chloranyl-2-fluoranyl-phenyl)carbamoylamino]-4-[cyclohexyl(2-methylpropyl)amino]phenyl]pyrrole-2-carboxylic acid, Indoleamine 2,3-dioxygenase 1, THIOSULFATE | | Authors: | Li, K, Liu, W, Dong, X. | | Deposit date: | 2023-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Apo-Form Selective Inhibition of IDO for Tumor Immunotherapy.
J Immunol., 209, 2022
|
|
6KFI
 
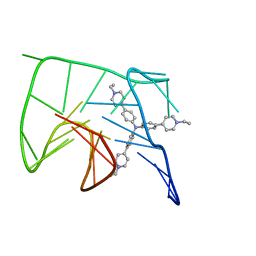 | | NMR solution structure of the 1:1 complex of Tel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA (26-MER) | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
