6MTJ
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies 3H109L and 35O22 at 2.9 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-19 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6MUF
 
 | |
6MTN
 
 | |
6MU8
 
 | |
6MU6
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-814508 in Complex with Human Antibodies 3H109L and 35O22 at 3.2 Angstrom | | Descriptor: | (2R)-{1-[{7-[2-({[3-(dimethylamino)propyl](methyl)amino}methyl)-1,3-thiazol-4-yl]-4-methoxy-1H-pyrrolo[2,3-c]pyridin-3-yl}(oxo)acetyl]piperidin-4-yl}(phenyl)acetonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
8X1N
 
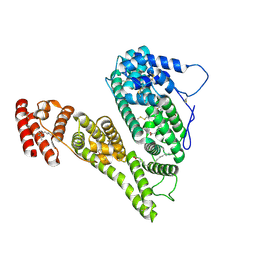 | | Cryo-EM structure of human alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein, PALMITIC ACID, ZINC ION, ... | | Authors: | Liu, Z.M, Li, M.S, Wu, C, Liu, K. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural characteristics of alpha-fetoprotein, including N-glycosylation, metal ion and fatty acid binding sites.
Commun Biol, 7, 2024
|
|
6EGC
 
 | |
3VDD
 
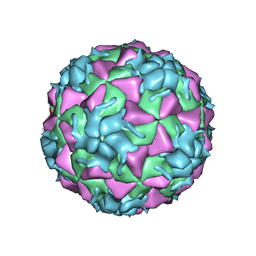 | | Structure of HRV2 capsid complexed with antiviral compound BTA798 | | Descriptor: | 3-ethoxy-6-{2-[1-(6-methylpyridazin-3-yl)piperidin-4-yl]ethoxy}-1,2-benzoxazole, Protein VP1, Protein VP2, ... | | Authors: | Morton, C.J, Feil, S.C, Parker, M.W. | | Deposit date: | 2012-01-05 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An Orally Available 3-Ethoxybenzisoxazole Capsid Binder with Clinical Activity against Human Rhinovirus.
ACS Med Chem Lett, 3, 2012
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
8TXG
 
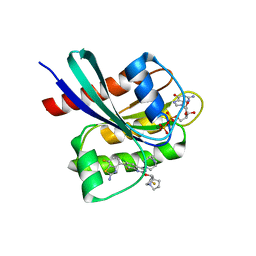 | | Crystal structure of KRAS G12D in complex with GDP and compound 8 | | Descriptor: | (4M)-4-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-7-fluoro-1,3-benzothiazol-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8TXH
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 14 | | Descriptor: | (4P)-2-amino-4-{4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}-6-(trifluoromethyl)quinazolin-7-yl}-7-fluoro-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8TXE
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 5 | | Descriptor: | (6M)-6-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-4-methyl-5-(trifluoromethyl)pyridin-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
7PRV
 
 | | The glucocorticoid receptor in complex with fluticasone furoate, a PGC1a coactivator fragment and sgk 23bp | | Descriptor: | (6alpha,11alpha,14beta,16alpha,17alpha)-6,9-difluoro-17-{[(fluoromethyl)sulfanyl]carbonyl}-11-hydroxy-16-methyl-3-oxoan drosta-1,4-dien-17-yl furan-2-carboxylate, 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*GP*TP*CP*GP*A)-3'), ... | | Authors: | Postel, S, Edman, K, Wissler, L. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PRW
 
 | | The glucocorticoid receptor in complex with velsecorat, a PGC1a coactivator fragment and sgk 23bp | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*AP*CP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*GP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*CP*GP*GP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*TP*GP*TP*AP*C)-3'), ... | | Authors: | Postel, S, Edman, K, Wissler, L. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PRX
 
 | | wildtype ligand binding domain of the glucocorticoid receptor complexed with velsecorat and a PGC1a coactivator fragment | | Descriptor: | Glucocorticoid receptor, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Velsecorat | | Authors: | Edman, K, Wissler, L, Koehler, C, Postel, S. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5XVF
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | Descriptor: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
4XIU
 
 | |
6NPW
 
 | | SSu72/Sympk in complex with Ser2/Ser5 phosphorylated peptide | | Descriptor: | PHOSPHATE ION, Ser2/Ser5 phosphorylated peptide, Ssu72 ortholog, ... | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Structural determinants for accurate dephosphorylation of RNA polymerase II by its cognate C-terminal domain (CTD) phosphatase during eukaryotic transcription.
J.Biol.Chem., 294, 2019
|
|
6G71
 
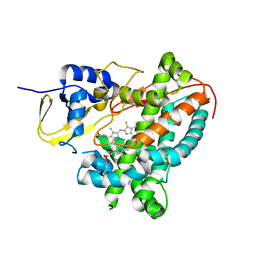 | | Structure of CYP1232A24 from Arthrobacter sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, FE (III) ION, ... | | Authors: | Dubiel, P, Sharma, M, Klenk, J, Hauer, B, Grogan, G. | | Deposit date: | 2018-04-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of cytochrome P450 1232A24 and 1232F1 from Arthrobacter sp. and their role in the metabolic pathway of papaverine.
J.Biochem., 166, 2019
|
|
6UBI
 
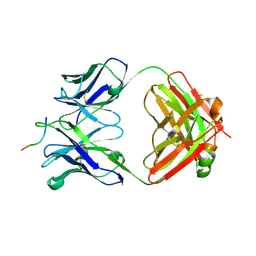 | |
6UCE
 
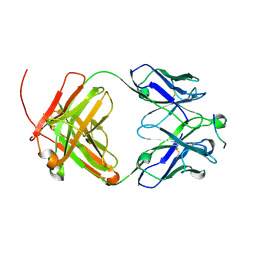 | |
6UCF
 
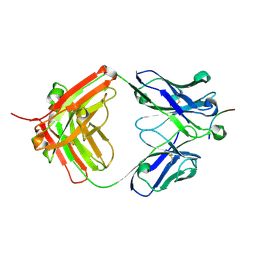 | |
6VRW
 
 | | Cryo-EM structure of stabilized HIV-1 Env trimer CAP256.wk34.c80 SOSIP.RnS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-03-11 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structure of Super-Potent Antibody CAP256-VRC26.25 in Complex with HIV-1 Envelope Reveals a Combined Mode of Trimer-Apex Recognition.
Cell Rep, 31, 2020
|
|
6VTT
 
 | |
