2VVV
 
 | | Aminopyrrolidine-related triazole Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-1H-1,2,4-triazol-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWO
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3S,4S)-4-FLUORO- 1-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWM
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | (4R)-4-{[(5-chlorothiophen-2-yl)carbonyl]amino}-N-(cyclopropylmethyl)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-L-prolinamide, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VVC
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[(3S,4S)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-4-methoxypyrrolidin-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-05 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWN
 
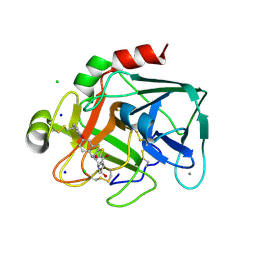 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid ((3S,4S)-1-{[2-fluoro-4-(2-oxo-2H-pyridin-1-yl)-phenylcarbamoyl]-methyl}-4-hydroxy-pyrrolidin-3-yl)-amide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
1MJ1
 
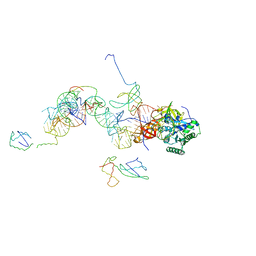 | | FITTING THE TERNARY COMPLEX OF EF-Tu/tRNA/GTP AND RIBOSOMAL PROTEINS INTO A 13 A CRYO-EM MAP OF THE COLI 70S RIBOSOME | | Descriptor: | Elongation Factor Tu, L11 ribosomal protein, Phe-tRNA, ... | | Authors: | Stark, H, Rodnina, M.V, Wieden, H.-J, Zemlin, F, Wintermeyer, W, Vanheel, M. | | Deposit date: | 2002-08-26 | | Release date: | 2002-11-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Ribosome Interactions of Aminoacyl-tRNA and Elongation Factor TU in the
Codon Recognition Complex
Nat.Struct.Biol., 9, 2002
|
|
1QIB
 
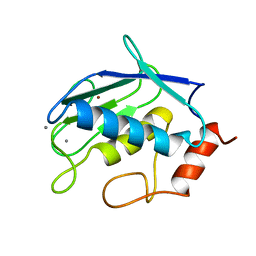 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
1BRD
 
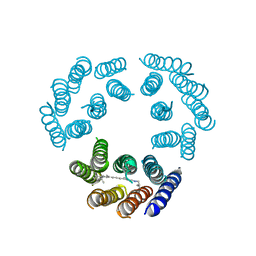 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
8SZM
 
 | |
8SZN
 
 | |
8FV4
 
 | | EGFR(T790M/V948R) in complex with compound 2 (LN5993) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(3P)-3-[(4P)-4-(2-acetamidopyridin-4-yl)-2-(methylsulfanyl)-1H-imidazol-5-yl]phenyl}-11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepine-9-carboxamide, ... | | Authors: | Ogboo, B.C, Beyett, T.S, Eck, M.J, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
8FV3
 
 | | EGFR(T790M/V948R) in complex with compound 1 (LN4503) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(4P)-4-[(4P)-5-{3-[(8-fluoro-11-oxo-5,11-dihydro-10H-dibenzo[b,e][1,4]diazepin-10-yl)methyl]phenyl}-2-(methylsulfanyl)-1H-imidazol-4-yl]pyridin-2-yl}acetamide, ... | | Authors: | Ogboo, B.C, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
6WA2
 
 | | Crystal structure of EGFR(T790M/V948R) in complex with LN3753 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-fluoro-5-hydroxybenzamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
6WAK
 
 | | A crystal structure of EGFR(T790M/V948R) in complex with LN3754 | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]benzamide, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
6WXN
 
 | | EGFR(T790M/V948R) in complex with LN3844 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
8TW1
 
 | | Crystal structure of Lys2972, a phage endolysin targeting Streptococcus thermophilus | | Descriptor: | Endolysin Lys2972, GLYCEROL, SODIUM ION | | Authors: | Zhu, X, Moineau, S, Shi, R. | | Deposit date: | 2023-08-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Fermentation Practices Select for Thermostable Endolysins in Phages.
Mol.Biol.Evol., 41, 2024
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
7R6H
 
 | | RHCC in complex with o-carborane | | Descriptor: | Tetrabrachion, ortho-carborane | | Authors: | Heide, F, McDougall, M, Stetefeld, J. | | Deposit date: | 2021-06-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Boron rich nanotube drug carrier system is suited for boron neutron capture therapy.
Sci Rep, 11, 2021
|
|
7MBI
 
 | | Structure of SARS-CoV2 3CL protease covalently bound to peptidomimetic inhibitor | | Descriptor: | 2,4,6-trimethylpyridine-3-carboxylic acid, 3C-like proteinase, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic alpha-Acyloxymethylketone Warheads with Six-Membered Lactam P1 Glutamine Mimic: SARS-CoV-2 3CL Protease Inhibition, Coronavirus Antiviral Activity, and in Vitro Biological Stability.
J.Med.Chem., 65, 2022
|
|
9ETC
 
 | |
9ETG
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with CA-M11 | | Descriptor: | Fatty acid-binding protein, liver, [4-[(2-azanyl-4-oxidanylidene-1,3-thiazol-5-yl)methyl]phenyl] (4~{R})-4-[(3~{R},5~{R},7~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoate | | Authors: | Tassone, G, Pozzi, C, Maramai, S. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETD
 
 | |
9ETE
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETF
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
5IDZ
 
 | |
