4DSZ
 
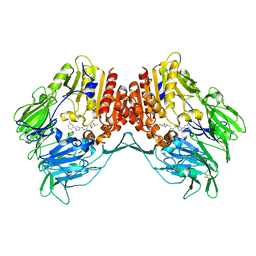 | | Crystal Structure of DPP-IV with Compound C2 | | Descriptor: | (2R)-4-[4-(3-methylphenyl)-1H-1,2,3-triazol-1-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine, Dipeptidyl peptidase 4 | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2012-02-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of DPP-IV with Compound C2
To be Published
|
|
4CMP
 
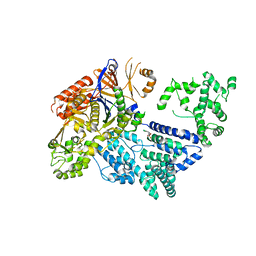 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
4DSA
 
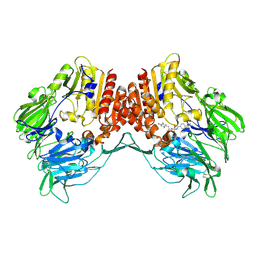 | | Crystal Structure of DPP-IV with Compound C1 | | Descriptor: | 4-[({[(2R)-2-amino-3-(2,4,5-trifluorophenyl)propyl]sulfamoyl}amino)methyl]benzenesulfonamide, Dipeptidyl peptidase 4 | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2012-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structure of DPP-IV with Compound C1
To be Published
|
|
5GGZ
 
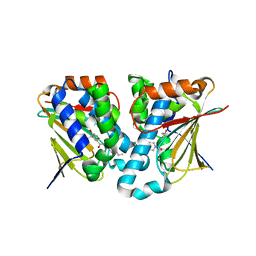 | | Crystal structure of novel inhibitor bound with Hsp90 | | Descriptor: | Heat shock protein HSP 90-alpha, [2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-(2-ethoxy-7,8-dihydro-5~{H}-pyrido[4,3-d]pyrimidin-6-yl)methanone | | Authors: | Chen, T.T, Li, J, Xu, Y.C. | | Deposit date: | 2016-06-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90
J. Med. Chem., 59, 2016
|
|
3MON
 
 | |
8WRH
 
 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRL
 
 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRM
 
 | | XBB.1.5 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
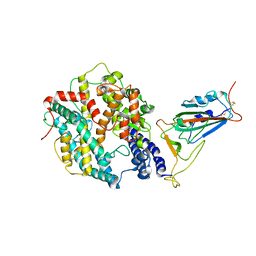 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRO
 
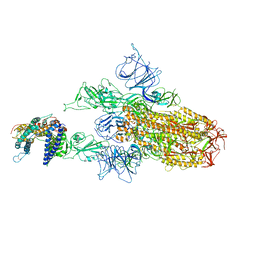 | | XBB.1.5.10 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
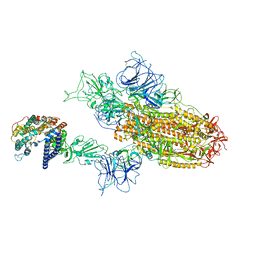 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
7YTO
 
 | |
5ZVL
 
 | | Crystal Structure of Wheat Glutarredoxin | | Descriptor: | Glutaredoxin | | Authors: | Hu, S.Q, Sun, X.M, Chen, M.R. | | Deposit date: | 2018-05-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Crystal Structure of Wheat Glutaredoxin and Its Application in Improving the Processing Quality of Flour.
J. Agric. Food Chem., 66, 2018
|
|
1J4J
 
 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
4ZZJ
 
 | | SIRT1/Activator/Substrate Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, Ac-p53, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7403 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4ZZH
 
 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4ZZI
 
 | | SIRT1/Activator/Inhibitor Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7346 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
7R8V
 
 | | Cryo-EM structure of the ADP state actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Espinosa de los Reyes, S, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-27 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB9
 
 | | Cryo-EM structure of the rigor state Jordan myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7R91
 
 | | cryo-EM structure of the rigor state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB8
 
 | | cryo-EM structure of the ADP state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7X31
 
 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7X4B
 
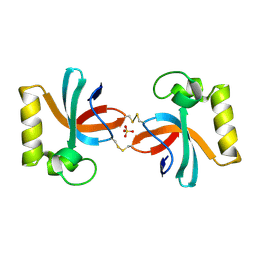 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
7C53
 
 | |
