1A23
 
 | | SOLUTION NMR STRUCTURE OF REDUCED DSBA FROM ESCHERICHIA COLI, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DSBA | | Authors: | Schirra, H.J, Renner, C, Czisch, M, Huber-Wunderlich, M, Holak, T.A, Glockshuber, R. | | Deposit date: | 1998-01-15 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of reduced DsbA from Escherichia coli in solution.
Biochemistry, 37, 1998
|
|
1A24
 
 | | SOLUTION NMR STRUCTURE OF REDUCED DSBA FROM ESCHERICHIA COLI, FAMILY OF 20 STRUCTURES | | Descriptor: | DSBA | | Authors: | Schirra, H.J, Renner, C, Czisch, M, Huber-Wunderlich, M, Holak, T.A, Glockshuber, R. | | Deposit date: | 1998-01-15 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of reduced DsbA from Escherichia coli in solution.
Biochemistry, 37, 1998
|
|
6T41
 
 | | CDK8/Cyclin C in complex with N-(4-chlorobenzyl)isoquinolin-4-amine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Schneider, E.V, Maskos, K, Huber, R, Kuhn, C.-D. | | Deposit date: | 2019-10-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A precisely positioned MED12 activation helix stimulates CDK8 kinase activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1TYX
 
 | | TITLE OF TAILSPIKE-PROTEIN | | Descriptor: | TAILSPIKE PROTEIN, alpha-D-galactopyranose-(1-2)-[alpha-D-Abequopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-alpha-D-galactopyranose-(1-2)-[alpha-D-Abequopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Steinbacher, S, Huber, R. | | Deposit date: | 1996-07-26 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1TYU
 
 | | STRUCTURE OF TAILSPIKE-PROTEIN | | Descriptor: | TAILSPIKE PROTEIN, alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Steinbacher, S, Huber, R. | | Deposit date: | 1996-07-26 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5JDW
 
 | |
2IXS
 
 | | Structure of SdaI restriction endonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SDAI RESTRICTION ENDONUCLEASE, ... | | Authors: | Tamulaitiene, G, Jakubauskas, A, Urbanke, C, Huber, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Rare-Cutting Restriction Enzyme Sdai Reveals Unexpected Domain Architecture
Structure, 14, 2006
|
|
4CTS
 
 | |
7A3F
 
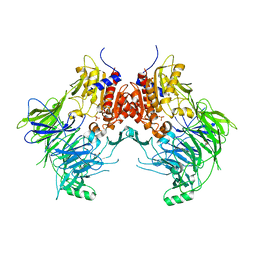 | | Crystal structure of apo DPP9 | | Descriptor: | Dipeptidyl peptidase 9, GLYCEROL, PHOSPHATE ION | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Development of 4-Oxo-beta-Lactams as Novel Inhibitors of Dipeptidyl Peptidases 8 and 9
To Be Published
|
|
7A3J
 
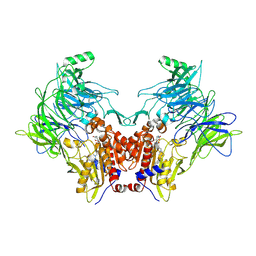 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, A272 | | Descriptor: | 2-ethyl-2-methanoyl-~{N}-[3-[[4-(naphthalen-1-ylmethyl)piperazin-1-yl]methyl]phenyl]butanamide, CHLORIDE ION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 2022
|
|
7A3G
 
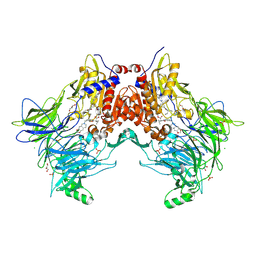 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, 91 | | Descriptor: | 1-[3-(7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-6-ylmethyl)phenyl]-3,3-diethyl-azetidine-2,4-dione, CHLORIDE ION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 2022
|
|
7A3I
 
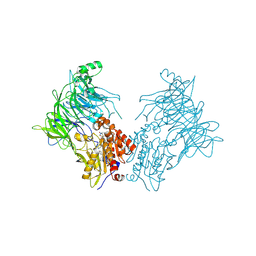 | |
7A3L
 
 | |
7A3K
 
 | |
7AYR
 
 | |
9JDW
 
 | |
7AYQ
 
 | |
2JDW
 
 | |
1TOC
 
 | |
2IMM
 
 | |
4AZU
 
 | |
1U9L
 
 | | Structural basis for a NusA- protein N interaction | | Descriptor: | GOLD ION, Lambda N, Transcription elongation protein nusA | | Authors: | Bonin, I, Muehlberger, R, Bourenkov, G.P, Huber, R, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-08-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6Z6F
 
 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
