4S3D
 
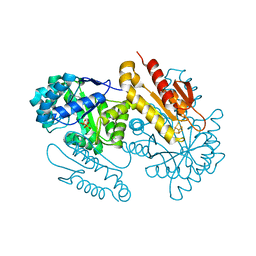 | | IspG in complex with PPi | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
9ASB
 
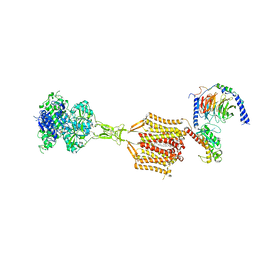 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
6OCP
 
 | | Crystal structure of a human GABAB receptor peptide bound to KCTD16 T1 | | Descriptor: | BTB/POZ domain-containing protein KCTD16, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OCT
 
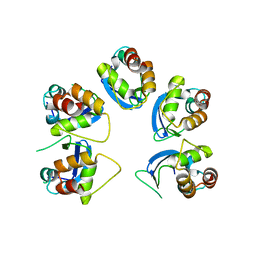 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OCR
 
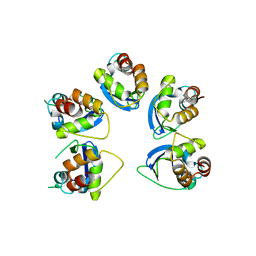 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6YS3
 
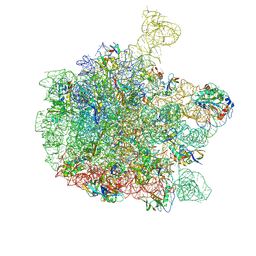 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
3ZI8
 
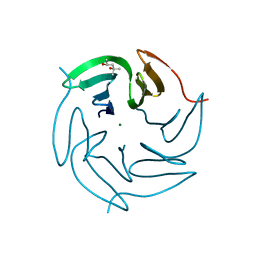 | | Structure of the R17A mutant of the Ralstonia soleanacerum lectin at 1.5 Angstrom in complex with L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, MAGNESIUM ION, PUTATIVE FUCOSE-BINDING LECTIN PROTEIN, ... | | Authors: | Arnaud, J, Audfray, A, Claudinon, J, Trondle, K, Trosvalet, M, Thomas, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reduction of Lectin Valency Drastically Changes Glycolipid Dynamics in Membranes, But not Surface Avidity.
Acs Chem.Biol., 8, 2013
|
|
4CSD
 
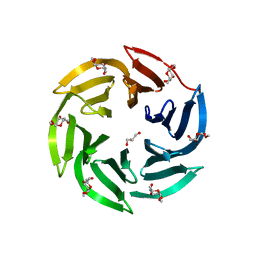 | | Structure of Monomeric Ralstonia solanacearum lectin | | Descriptor: | FUCOSE-BINDING LECTIN PROTEIN, GLYCEROL, methyl alpha-L-fucopyranoside | | Authors: | Arnaud, J, Trundle, K, Claudinon, J, Audfray, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2014-03-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Membrane Deformation by Neolectins with Engineered Glycolipid Binding Sites.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5A3O
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa in complex with Methyl 6-(cinnamido)-6-deoxy-alpha-D-mannopyranoside at 1.6 ansgtrom | | Descriptor: | CALCIUM ION, CHLORIDE ION, CINNAMIDE, ... | | Authors: | Sommer, R, Hauck, D, Varrot, A, Audfray, A, Imberty, A, Titz, A. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cinnamide Derivatives of D-Mannose as Inhibitors of the Bacterial Virulence Factor Lecb from Pseudomonas Aeruginosa
Chemistryopen, 4, 2015
|
|
3ZHB
 
 | |
3ZGY
 
 | |
4CPD
 
 | | Alcohol dehydrogenase TADH from Thermus sp. ATN1 | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Man, H, Gargulio, S, Frank, A, Hollmann, F, Grogan, G. | | Deposit date: | 2014-02-05 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Nadh-Dependent Thermostable Alcohol Dehydrogenase Tadh from Thermus Sp. Atn1 Provides a Platform for Engineering Specificity and Improved Compatibility with Inorganic Cofactor-Regeneration Catalysts
J.Mol.Catal., B Enzym., 105, 2014
|
|
4CY8
 
 | | 2-hydroxybiphenyl 3-monooxygenase (HbpA) in complex with FAD | | Descriptor: | 2-HYDROXYBIPHENYL 3-MONOOXYGENASE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Jensen, C.N, Farrugia, J.E, Frank, A, Man, H, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2014-04-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of the Apo and Fad-Bound Forms of 2-Hydroxybiphenyl 3-Monooxygenase (Hbpa) Locate Activity Hotspots Identified by Using Directed Evolution.
Chembiochem, 16, 2015
|
|
5MRN
 
 | | Arabidopsis thaliana IspD Glu258Ala Mutant | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRP
 
 | | Arabidopsis thaliana IspD Glu258Ala mutant in complex with Azolopyrimidine (2) | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, 5-chloro-7-hydroxy-6-(phenylmethyl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRQ
 
 | | Arabidopsis thaliana IspD Asp262Ala Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRM
 
 | | Arabidopsis thaliana IspD in complex with Isoxazole (4) | | Descriptor: | 2,4-bis(bromanyl)-6-[3-(trifluoromethyl)-1,2-oxazol-5-yl]phenol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRO
 
 | | Arabidopsis thaliana IspD Glu258Ala mutant in complex with Azolopyrimidine (1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
6BX9
 
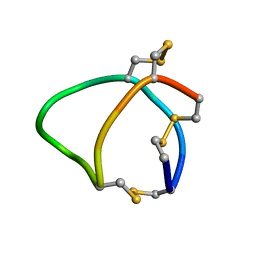 | | Solution structure of conotoxin reg3b | | Descriptor: | Conotoxin | | Authors: | Mari, F, Dovell, S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural plasticity of mini-M conotoxins - expression of all mini-M subtypes by Conus regius.
FEBS J., 285, 2018
|
|
7B8Q
 
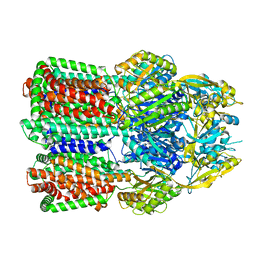 | | Acinetobacter baumannii multidrug transporter AdeB in L*OO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8P
 
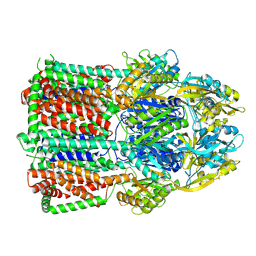 | | Acinetobacter baumannii multidrug transporter AdeB in OOO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
8Q91
 
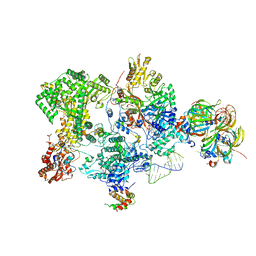 | | Structure of the human 20S U5 snRNP core | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4U6H
 
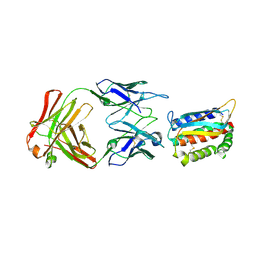 | | Vaccinia L1/M12B9-Fab complex | | Descriptor: | Heavy chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Light chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Protein L1 | | Authors: | Matho, M.H, Schlossman, A, Zajonc, D.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent neutralization of vaccinia virus by divergent murine antibodies targeting a common site of vulnerability in l1 protein.
J.Virol., 88, 2014
|
|
7QUH
 
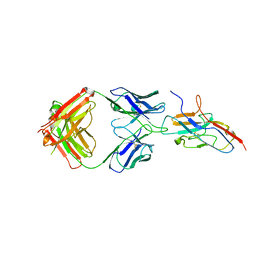 | | Siglec-8 in complex with therapeutic Fab AK002. | | Descriptor: | Sialic acid-binding Ig-like lectin 8, Sialic acid-binding immunoglobulin-type lectin | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez Barbero, J, Ereno Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
9FA7
 
 | |
