1DNP
 
 | | STRUCTURE OF DEOXYRIBODIPYRIMIDINE PHOTOLYASE | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Park, H.-W, Sancar, A, Deisenhofer, J. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of DNA photolyase from Escherichia coli.
Science, 268, 1995
|
|
1HR6
 
 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HWJ
 
 | |
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HW8
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH COMPACTIN (ALSO KNOWN AS MEVASTATIN) | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,8S,8aR)-2-methyl-8-{[(2S)-2-methylbutanoyl]oxy}-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]h eptanoic acid, ADENOSINE-5'-DIPHOSPHATE, HMG-COA REDUCTASE | | Authors: | Istvan, E.S, Deisenhofer, J. | | Deposit date: | 2001-01-09 | | Release date: | 2001-05-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural mechanism for statin inhibition of HMG-CoA reductase.
Science, 292, 2001
|
|
1HWL
 
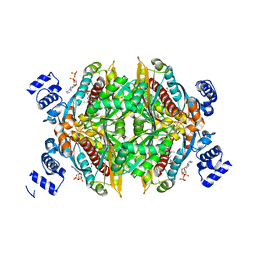 | |
1N7D
 
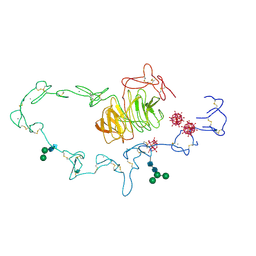 | | Extracellular domain of the LDL receptor | | Descriptor: | 12-TUNGSTOPHOSPHATE, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Rudenko, G, Henry, L, Henderson, K, Ichtchenko, K, Brown, M.S, Goldstein, J.L, Deisenhofer, J. | | Deposit date: | 2002-11-13 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the LDL receptor extracellular domain at endosomal pH
Science, 298, 2002
|
|
1I7N
 
 | |
1I7L
 
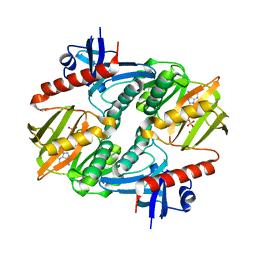 | |
1KMP
 
 | | Crystal structure of the Outer Membrane Transporter FecA Complexed with Ferric Citrate | | Descriptor: | CITRIC ACID, FE (III) ION, IRON(III) DICITRATE TRANSPORT PROTEIN FECA, ... | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1KMO
 
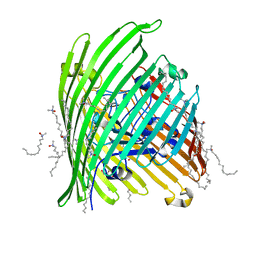 | | Crystal structure of the Outer Membrane Transporter FecA | | Descriptor: | HEPTANE-1,2,3-TRIOL, Iron(III) dicitrate transport protein fecA, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1MZE
 
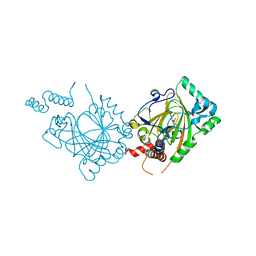 | | Human Factor Inhibiting HIF (FIH1) | | Descriptor: | D(-)-TARTARIC ACID, FE (II) ION, factor inhibiting HIF1 | | Authors: | Dann III, C.E, Bruick, R.K, Deisenhofer, J. | | Deposit date: | 2002-10-07 | | Release date: | 2002-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor 1: An Asparaginyl Hydroxylase Involved in the Hypoxic Response Pathway
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MZF
 
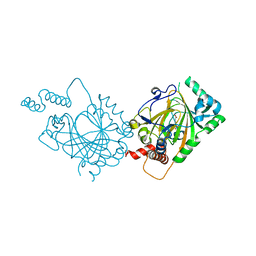 | |
1MHC
 
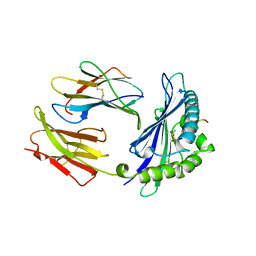 | | MODEL OF MHC CLASS I H2-M3 WITH NONAPEPTIDE FROM RAT ND1 REFINED AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I ANTIGEN H2-M3, NONAPEPTIDE FROM RAT NADH DEHYDROGENASE | | Authors: | Wang, C.-R, Fischer Lindahl, K, Deisenhofer, J. | | Deposit date: | 1995-08-23 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3
Cell(Cambridge,Mass.), 82, 1995
|
|
1BIF
 
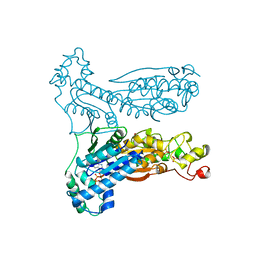 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE BIFUNCTIONAL ENZYME COMPLEXED WITH ATP-G-S AND PHOSPHATE | | Descriptor: | 6-PHOSPHOFRUCTO-2-KINASE/ FRUCTOSE-2,6-BISPHOSPHATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hasemann, C.A, Deisenhofer, J. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies.
Structure, 4, 1996
|
|
1C4O
 
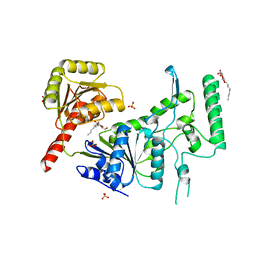 | | CRYSTAL STRUCTURE OF THE DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB FROM THERMUS THERMOPHILUS | | Descriptor: | DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Machius, M, Henry, L, Palnitkar, M, Deisenhofer, J. | | Deposit date: | 1999-09-14 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C4R
 
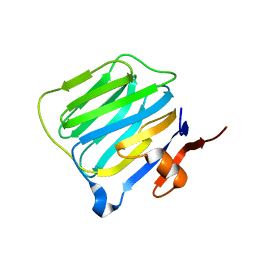 | | THE STRUCTURE OF THE LIGAND-BINDING DOMAIN OF NEUREXIN 1BETA: REGULATION OF LNS DOMAIN FUNCTION BY ALTERNATIVE SPLICING | | Descriptor: | NEUREXIN-I BETA | | Authors: | Rudenko, G, Nguyen, T, Chelliah, Y, Sudhof, T.C, Deisenhofer, J. | | Deposit date: | 1999-09-28 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing.
Cell(Cambridge,Mass.), 99, 1999
|
|
1U3D
 
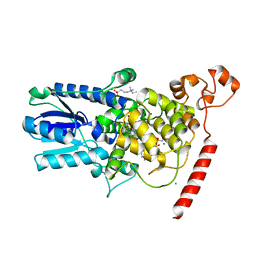 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S2J
 
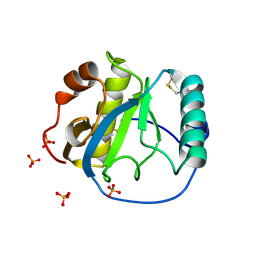 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
1U3C
 
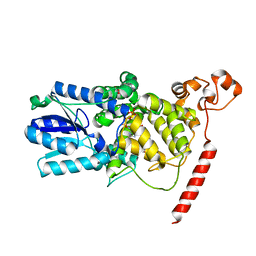 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1DQ8
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG AND COA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1DQ9
 
 | | COMPLEX OF CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG-COA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, PROTEIN (HMG-COA REDUCTASE) | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1DQA
 
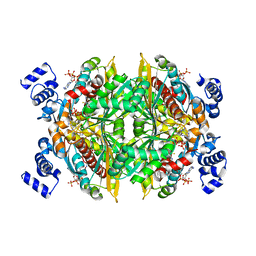 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG, COA, AND NADP+ | | Descriptor: | 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1H5B
 
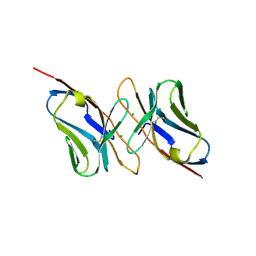 | | T cell receptor Valpha11 (AV11S5) domain | | Descriptor: | CHLORIDE ION, GLYCEROL, MURINE T CELL RECEPTOR (TCR) VALPHA DOMAIN | | Authors: | Machius, M, Cianga, P, Deisenhofer, J, Sally Ward, E. | | Deposit date: | 2001-05-21 | | Release date: | 2001-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a T Cell Receptor Valpha11 (Av11S5) Domain: New Canonical Forms for the First and Second Complementarity Determining Regions
J.Mol.Biol., 310, 2001
|
|
1FEP
 
 | | FERRIC ENTEROBACTIN RECEPTOR | | Descriptor: | FERRIC ENTEROBACTIN RECEPTOR | | Authors: | Buchanan, S.K, Smith, B.S, Ventatramani, L, Xia, D, Esser, L, Palnitkar, M, Chakraborty, R, Van Der Helm, D, Deisenhofer, J. | | Deposit date: | 1998-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the outer membrane active transporter FepA from Escherichia coli.
Nat.Struct.Biol., 6, 1999
|
|
