1YEM
 
 | | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein Pfu-838710-001, PLATINUM (II) ION, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chang, J, Shah, A, Ng, J.D, Liu, Z.-J, Chen, L, Lee, D, Tempel, W, Praissman, J.L, Lin, D, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus
To be published
|
|
2VER
 
 | | Structural model for the complex between the Dr adhesins and carcinoembryonic antigen (CEA) | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, ARCINOEMBRYONIC ANTIGEN-RELATED CELL ADHESION MOLECULE 5, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Korotkova, N, Yang, Y, Le Trong, I, Cota, E, Demeler, B, Marchant, J, Thomas, W.E, Stenkamp, R.E, Moseley, S.L, Matthews, S. | | Deposit date: | 2007-10-26 | | Release date: | 2008-01-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Binding of Dr Adhesins of Escherichia Coli to Carcinoembryonic Antigen Triggers Receptor Dissociation.
Mol.Microbiol., 67, 2008
|
|
2HQ4
 
 | | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH1570 | | Authors: | Li, Y, Marshall, M, Chang, J, Zhao, M, Zhang, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii
To be Published
|
|
2IA0
 
 | | Transcriptional Regulatory Protein PF0864 From Pyrococcus Furiosus a Member of the ASNC Family (PF0864) | | Descriptor: | GOLD ION, Putative HTH-type transcriptional regulator PF0864 | | Authors: | Yang, H, Chang, J, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structure of the Transcriptional Regulatory Protein PF0864: an Asnc Family member from Pyrococcus Furiosus
To be Published
|
|
1ABV
 
 | | N-TERMINAL DOMAIN OF THE DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE FROM ESCHERICHIA COLI, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE | | Authors: | Wilkens, S, Dunn, S.D, Chandler, J, Dahlquist, F.W, Capaldi, R.A. | | Deposit date: | 1997-01-29 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of the delta subunit of the E. coli ATPsynthase.
Nat.Struct.Biol., 4, 1997
|
|
4EMN
 
 | | Crystal structure of RpfB catalytic domain in complex with benzamidine | | Descriptor: | BENZAMIDINE, Probable resuscitation-promoting factor rpfB, SULFATE ION | | Authors: | Ruggiero, A, Marchant, J, Squeglia, F, Makarov, V, De Simone, A, Berisio, R. | | Deposit date: | 2012-04-12 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular determinants of inactivation of the resuscitation promoting factor B from Mycobacterium tuberculosis.
J.Biomol.Struct.Dyn., 31, 2013
|
|
4DM9
 
 | | The Crystal Structure of Ubiquitin Carboxy-terminal hydrolase L1 (UCHL1) bound to a tripeptide fluoromethyl ketone Z-VAE(OMe)-FMK | | Descriptor: | Tripeptide fluoromethyl ketone inhibitor Z-VAE(OMe)-FMK, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Davies, C.W, Chaney, J, Korbel, G, Ringe, D, Petsko, G.A, Ploegh, H, Das, C. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK).
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2MP2
 
 | | Solution structure of SUMO dimer in complex with SIM2-3 from RNF4 | | Descriptor: | E3 ubiquitin-protein ligase RNF4, Small ubiquitin-related modifier 3 | | Authors: | Xu, Y, Plechanovov, A, Simpson, P, Marchant, J, Leidecker, O, Sebastian, K, Hay, R.T, Matthews, S.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Nat Commun, 5, 2014
|
|
4CKB
 
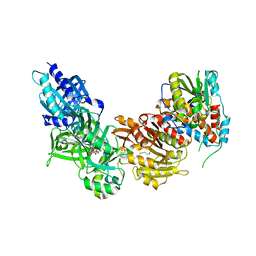 | | Vaccinia virus capping enzyme complexed with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, ... | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4CKE
 
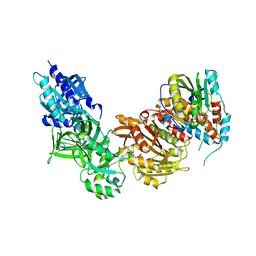 | | Vaccinia virus capping enzyme complexed with SAH in P1 form | | Descriptor: | MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
1RQ5
 
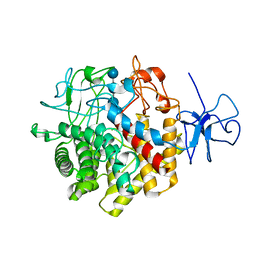 | | Structural Basis for the Exocellulase Activity of the Cellobiohydrolase CbhA from C. thermocellum | | Descriptor: | CALCIUM ION, Cellobiohydrolase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Schubot, F.D, Kataeva, I.A, Chang, J, Shah, A.K, Ljungdahl, L.G, Rose, J.P, Wang, B.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the exocellulase activity of the cellobiohydrolase CbhA from Clostridium thermocellum
Biochemistry, 43, 2004
|
|
2LU2
 
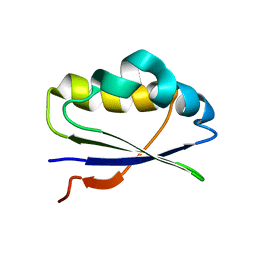 | | MIC5 regulates the activity of Toxoplasma subtilisin 1 by mimicking a subtilisin prodomain | | Descriptor: | Microneme TgMIC5 protein | | Authors: | Saouros, S, Dou, Z, Henry, M, Marchant, J, Carruthers, V.B, Matthews, S. | | Deposit date: | 2012-06-07 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Microneme protein 5 regulates the activity of toxoplasma subtilisin 1 by mimicking a subtilisin prodomain.
J.Biol.Chem., 287, 2012
|
|
2JEV
 
 | | Crystal structure of human spermine,spermidine acetyltransferase in complex with a bisubstrate analog (N1-acetylspermine-S-CoA). | | Descriptor: | (3R)-27-AMINO-3-HYDROXY-2,2-DIMETHYL-4,8,14-TRIOXO-12-THIA-5,9,15,19,24-PENTAAZAHEPTACOS-1-YL [(2S,3R,4S,5S)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, DIAMINE ACETYLTRANSFERASE 1 | | Authors: | Hegde, S.S, Chandler, J, Vetting, M.W, Yu, M, Blanchard, J.S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic and Structural Analysis of Human Spermidine/Spermine N(1)-Acetyltransferase.
Biochemistry, 46, 2007
|
|
2M4H
 
 | | Solution structure of the Core Domain (10-76) of the Feline Calicivirus VPg protein | | Descriptor: | Feline Calicivirus VPg protein | | Authors: | Kwok, R.N, Leen, E.N, Birtley, J.R, Prater, S.N, Simpson, P.J, Curry, S, Matthews, S, Marchant, J. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
2IDG
 
 | | Crystal Structure of hypothetical protein AF0160 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0160 | | Authors: | Zhao, M, Zhang, M, Chang, J, Chen, L, Xu, H, Li, Y, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Hypothetical Protein AF0160 from Archaeoglobus fulgidus at 2.69 Angstrom resolution
To be Published
|
|
2MPV
 
 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
2K2T
 
 | | Epidermal growth Factor-like domain 2 from Toxoplasma gondii Microneme protein 6 | | Descriptor: | Micronemal protein 6 | | Authors: | Sawmynaden, K, Saouros, S, Marchant, J, Simpson, P, Matthews, S. | | Deposit date: | 2008-04-11 | | Release date: | 2009-02-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural insights into microneme protein assembly reveal a new mode of EGF domain recognition.
Embo Rep., 9, 2008
|
|
2M4G
 
 | | Solution structure of the Core Domain (11-85) of the Murine Norovirus VPg protein | | Descriptor: | Murine Norovirus VPg protein | | Authors: | Leen, E.N, Kwok, R, Birtley, J.R, Prater, S.N, Simpson, P.J, Matthews, S, Marchant, J, Curry, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
7DEY
 
 | | Structure of Dicer from Pichia stipitis | | Descriptor: | RNase III | | Authors: | Jobichen, C, Jingru, C. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structural and mechanistic insight into stem-loop RNA processing by yeast Pichia stipitis Dicer.
Protein Sci., 30, 2021
|
|
1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2J
 
 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2N
 
 | |
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
6DGT
 
 | | Selective PI3K beta inhibitor bound to PI3K delta | | Descriptor: | 4-[1-(5,8-difluoroquinolin-4-yl)-2-methyl-4-(4H-1,2,4-triazol-3-yl)-1H-benzimidazol-6-yl]-3-fluoropyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J, Villasenor, A, McGrath, M. | | Deposit date: | 2018-05-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Atropisomerism by Design: Discovery of a Selective and Stable Phosphoinositide 3-Kinase (PI3K) beta Inhibitor.
J. Med. Chem., 61, 2018
|
|
