5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
5YZ0
 
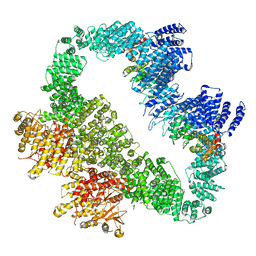 | | Cryo-EM Structure of human ATR-ATRIP complex | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | Authors: | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
5Y3R
 
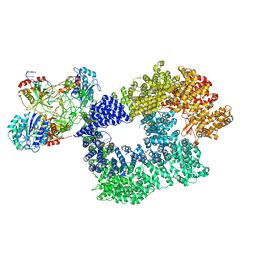 | | Cryo-EM structure of Human DNA-PK Holoenzyme | | Descriptor: | DNA (34-MER), DNA (36-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Yin, X, Liu, M, Tian, Y, Wang, J, Xu, Y. | | Deposit date: | 2017-07-29 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of human DNA-PK holoenzyme
Cell Res., 27, 2017
|
|
7S0N
 
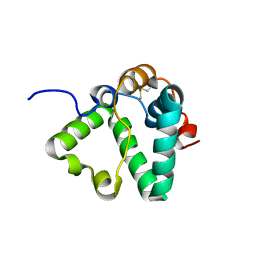 | | Structure of MS3494 from Mycobacterium Smegmatis determined by Solution NMR | | Descriptor: | Secreted protein | | Authors: | Kent, J.E, Tian, Y, Shin, K, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Method: | SOLUTION NMR | | Cite: | Structure of MS3494 from Mycobacterium Smegmatis
To Be Published
|
|
5AC3
 
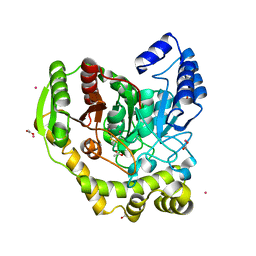 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
7FI9
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI7
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI8
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI6
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI5
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7TXR
 
 | |
7U68
 
 | |
2GX6
 
 | |
5HMH
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | 4-[2-(4-{[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]carbonyl}piperazin-1-yl)phenoxy]butanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HMK
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {4-[2-(2-hydroxyethoxy)phenyl]piperazin-1-yl}[(2R,3S)-2-propyl-3-[4-(trifluoromethyl)phenoxy]-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}piperidin-3-yl]methanone | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
7YVC
 
 | | Aplysia californica FaNaC in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
7YVB
 
 | | Aplysia californica FaNaC in ligand bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide-gated Na+ channel, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
7YLM
 
 | | Cryo-EM structure of 8-subunit Smc5/6 hinge region | | Descriptor: | MMS21 isoform 1, SMC6 isoform 1, Structural maintenance of chromosomes protein 5 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.17 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7YQH
 
 | | Cryo-EM structure of 8-subunit Smc5/6 | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 3, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2022-08-07 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7YMD
 
 | | Cryo-EM structure of Nse1/3/4 | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | Authors: | Qian, L, Jun, Z, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.176 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
5I05
 
 | |
6K9K
 
 | | Monomeric human ATM (Ataxia telangiectasia mutated) kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Yuriy, C, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
2LJ2
 
 | | Integral membrane core domain of the mercury transporter MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Das, B.B, Nothnagel, H.J, Lu, G.J, Son, W, Park, S, Tian, Y.B, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-09-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of a membrane protein in proteoliposomes.
J.Am.Chem.Soc., 134, 2012
|
|
5JZV
 
 | | The structure of D77G hCINAP-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase isoenzyme 6 | | Authors: | Liu, Y, Yang, Z, Yang, Y, Cai, X, Zheng, X. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumour growth.
Nat Commun, 7, 2016
|
|
4UWD
 
 | |
