6KNM
 
 | | Apelin receptor in complex with single domain antibody | | Descriptor: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | Authors: | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
1P5Q
 
 | | Crystal Structure of FKBP52 C-terminal Domain | | Descriptor: | FK506-binding protein 4, SULFATE ION | | Authors: | Wu, B, Li, P, Lou, Z, Shu, C, Ding, Y, Shen, B, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1N1A
 
 | | Crystal Structure of the N-terminal domain of human FKBP52 | | Descriptor: | FKBP52 | | Authors: | Li, P, Ding, Y, Wu, B, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-terminal domain of human FKBP52.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4GXO
 
 | | Crystal structure of Staphylococcus aureus protein SarZ mutant C13E | | Descriptor: | GLYCEROL, MarR family regulatory protein | | Authors: | Sun, F, Ding, Y, Ji, Q, Liang, Z, Deng, X, Wong, C.C, Yi, C, Zhang, L, Xie, S, Alvarez, S, Hicks, L.M, Luo, C, Jiang, H, Lan, L, He, C. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1QZ2
 
 | | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | | Descriptor: | 5-mer peptide from Heat shock protein HSP 90, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Liu, Y, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-09-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7XQV
 
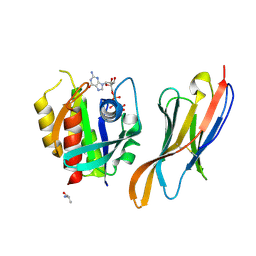 | | The complex of nanobody Rh57 binding to GTP-bound RhoA active form | | Descriptor: | ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhang, Y.R, Liu, R, Ding, Y. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insights into the binding of nanobody Rh57 to active RhoA-GTP.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
7V99
 
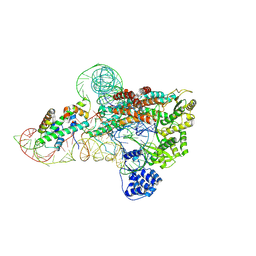 | | catalytic core of human telomerase holoenzyme | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Primer DNA, ... | | Authors: | Wan, F, Ding, Y, Yang, L, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Zipper head mechanism of telomere synthesis by human telomerase.
Cell Res., 31, 2021
|
|
7V9A
 
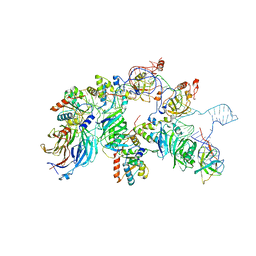 | | biogenesis module of human telomerase holoenzyme | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | Authors: | Wan, F, Ding, Y, Yang, L, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Zipper head mechanism of telomere synthesis by human telomerase.
Cell Res., 31, 2021
|
|
1Q1C
 
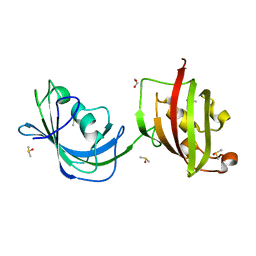 | | Crystal structure of N(1-260) of human FKBP52 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-07-18 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
2N2M
 
 | |
2LBW
 
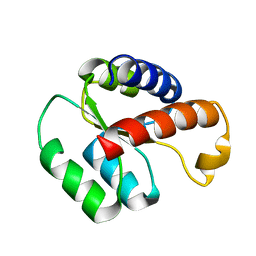 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p-S82W mutant | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
2LBX
 
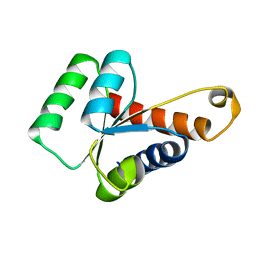 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
4QBA
 
 | |
7XOI
 
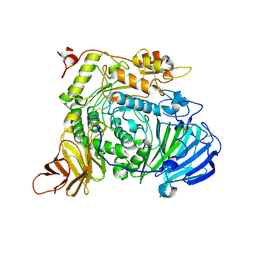 | | Aspergillus sojae alpha-glucosidase AsojAgdL in complex with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic processing of Aspergillus sojae alpha-glucosidase L with strong transglucosylation activity.
J.Struct.Biol., 214, 2022
|
|
7W8N
 
 | | Microbial Hormone-sensitive lipase E53 wild type | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-23 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
7CI0
 
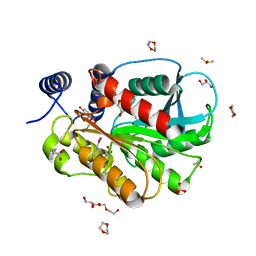 | | Microbial Hormone-sensitive lipase E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
7CIH
 
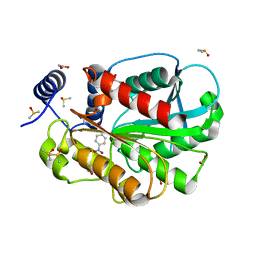 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
9KQK
 
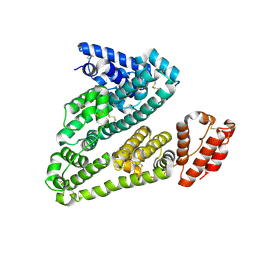 | | Crystal structure of HSA in complex with AmpHecy | | Descriptor: | 2-[[6-[5-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]thiophen-2-yl]naphthalen-2-yl]-methyl-amino]ethanol, Albumin | | Authors: | Chen, X, Shi, M.W, Ge, Y.H, Fang, B, Li, L. | | Deposit date: | 2024-11-26 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Fluorogen-Activating Human Serum Albumin for Mitochondrial Nanoscale Imaging.
Adv Mater, 2025
|
|
5JVA
 
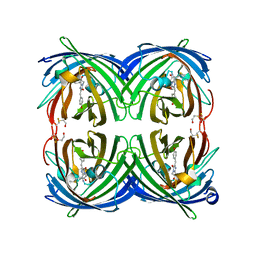 | | 1.95 angstrom crystal structure of TAGRFP-T | | Descriptor: | BETA-MERCAPTOETHANOL, SULFATE ION, TagRFP-T | | Authors: | Hu, X.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of red fluorescent protein TagRFP-T reveals the mechanism of its superior photostability.
Biochem. Biophys. Res. Commun., 477, 2016
|
|
5XM7
 
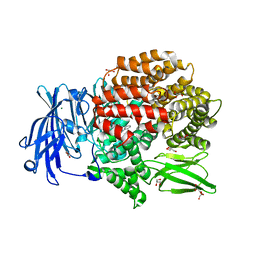 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-4-methyl-N-[(1R)-2-(oxidanylamino)-2-oxidanylidene-1-phenyl-ethyl]-2-[(phenylmethyl)carbamoylamino]pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-05-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y1T
 
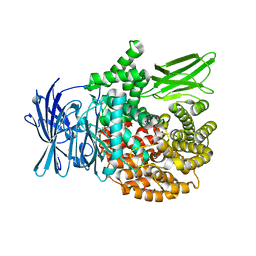 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,3-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(2,3-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
1UEF
 
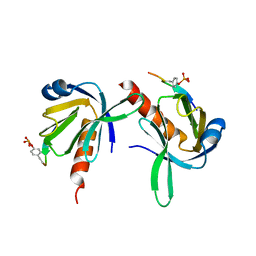 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
6XRT
 
 | |
