1XET
 
 | | Crystal structure of stilbene synthase from Pinus sylvestris, complexed with methylmalonyl CoA | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Dihydropinosylvin synthase, METHYLMALONYL-COENZYME A | | Authors: | Ng, S.H, Chirgadze, D, Spiteller, D, Li, T.L, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2004-09-12 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of stilbene synthase from Pinus sylvestris, complexed with methylmalonyl CoA
To be Published
|
|
1BLX
 
 | | P19INK4D/CDK6 COMPLEX | | Descriptor: | CALCIUM ION, CYCLIN-DEPENDENT KINASE 6, P19INK4D | | Authors: | Brotherton, D.H, Dhanaraj, V, Wick, S, Brizuela, L, Domaille, P.J, Volyanik, E, Xu, X, Parisini, E, Smith, B.O, Archer, S.J, Serrano, M, Brenner, S.L, Blundell, T.L, Laue, E.D. | | Deposit date: | 1998-07-21 | | Release date: | 1999-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d.
Nature, 395, 1998
|
|
2J66
 
 | | Structural characterisation of BtrK decarboxylase from butirosin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, BTRK, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Popovic, B, Li, Y, Chirgadze, D.Y, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterisation of Btrk Decarboxylase from Bacillus Circulans Butirosin Biosynthesis
To be Published
|
|
2JXR
 
 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
4GCR
 
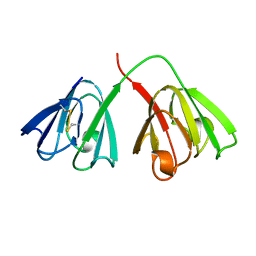 | | STRUCTURE OF THE BOVINE EYE LENS PROTEIN GAMMA-B (GAMMA-II)-CRYSTALLIN AT 1.47 ANGSTROMS | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Slingsby, C, Najmudin, S, Nalini, V, Driessen, H.P.C, Blundell, T.L, Moss, D.S, Lindley, P. | | Deposit date: | 1992-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the bovine eye lens protein gammaB(gammaII)-crystallin at 1.47 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
8OWP
 
 | |
8OWR
 
 | |
8OW5
 
 | |
8OWB
 
 | |
8OWQ
 
 | |
8QID
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with fragment | | Descriptor: | 1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QJ8
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 3-[3-(3-azanyl-2-cyano-phenyl)indol-1-yl]propanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QIX
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 3-(3-methylindol-1-yl)-~{N}-(4-phenoxyphenyl)sulfonyl-propanamide, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QIY
 
 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 1-(2-aminophenyl)-5-(trifluoromethyl)pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.5149 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
2Q1T
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmF in complex with NAD+ and UDP | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase, URIDINE-5'-DIPHOSPHATE | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2Q1W
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmH in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | King, J.D, Harmer, N.J, Maskell, D.J, Blundell, T.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PZM
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmG in complex with NAD and UDP | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase, SULFATE ION, ... | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PZK
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmG in complex with NAD | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | King, J.D, Harmer, N.J, Maskell, D.J, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2Q1U
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmF in complex with NAD+ and UDP | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase, ... | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PZL
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmG in complex with NAD and UDP | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase, URIDINE-5'-DIPHOSPHATE | | Authors: | King, J.D, Harmer, N.J, Maskell, D.J, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2Q1S
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmF in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PZJ
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmF in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
6ZH8
 
 | | Cryo-EM structure of DNA-PKcs:DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHA
 
 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3DAR
 
 | |
