8JMH
 
 | |
3BX5
 
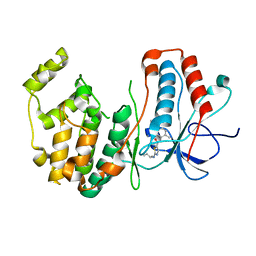 | | P38 alpha map kinase complexed with BMS-640994 | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-methyl-5-(methylcarbamoyl)phenyl]-2-{[(1R)-1-methylpropyl]amino}-1,3-thiazole-5-carboxamide | | Authors: | Sack, J.S. | | Deposit date: | 2008-01-11 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The discovery of (R)-2-(sec-butylamino)-N-(2-methyl-5-(methylcarbamoyl)phenyl) thiazole-5-carboxamide (BMS-640994)-A potent and efficacious p38alpha MAP kinase inhibitor
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4FFW
 
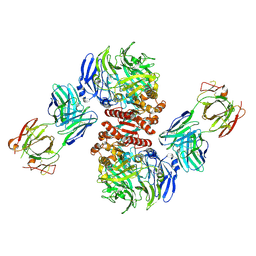 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
2ZGJ
 
 | | Crystal Structure of D86N-GzmM Complexed with Its Optimal Synthesized Substrate | | Descriptor: | Granzyme M, SSGKVPLS, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
5DO4
 
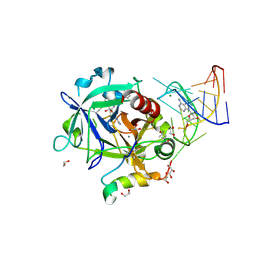 | | Thrombin-RNA aptamer complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Evoking picomolar binding in RNA by a single phosphorodithioate linkage.
Nucleic Acids Res., 44, 2016
|
|
7LRD
 
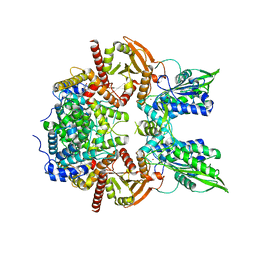 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LRE
 
 | |
7LRC
 
 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
8XSS
 
 | |
8XSR
 
 | |
8XST
 
 | |
7L28
 
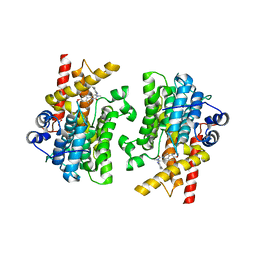 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | Descriptor: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-12-16 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L29
 
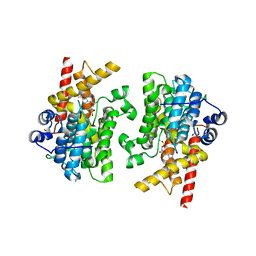 | |
7L27
 
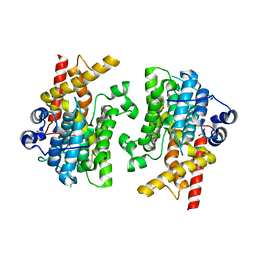 | |
8XK2
 
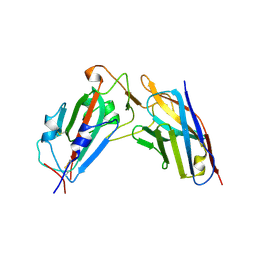 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | Spike protein S1, VHH60 nanobody | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8XKI
 
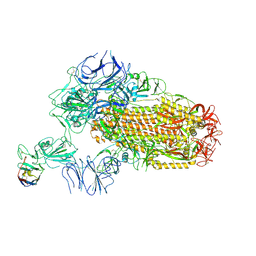 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
5AYW
 
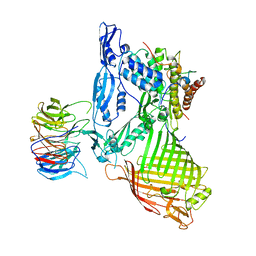 | | Structure of a membrane complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Huang, Y, Han, L, Zheng, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins
Nat.Struct.Mol.Biol., 23, 2016
|
|
5DK3
 
 | | Crystal Structure of Pembrolizumab, a full length IgG4 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain, ... | | Authors: | Scapin, G, Prosise, W, Reichert, P. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7KWE
 
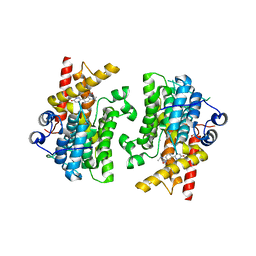 | | Crystal structure of the catalytic domain of human PDE3A bound to DNMDP | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7V2Z
 
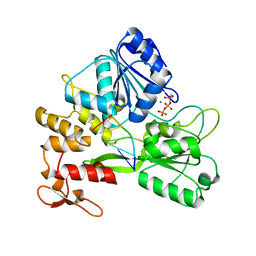 | | ZIKV NS3helicase in complex with ssRNA and ATP-Mn2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Core protein, MANGANESE (II) ION, ... | | Authors: | Lin, M.M, Yang, H.T. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10101676 Å) | | Cite: | Structural Basis of Zika Virus Helicase in RNA Unwinding and ATP Hydrolysis.
Acs Infect Dis., 8, 2022
|
|
7SNI
 
 | | Structure of G6PD-D200N tetramer bound to NADP+ and G6P | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TOF
 
 | | Structure of G6PD-WT dimer with no symmetry applied | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TOE
 
 | |
7SNG
 
 | | structure of G6PD-WT tetramer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SNH
 
 | | Structure of G6PD-D200N tetramer bound to NADP+ | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
