4QUU
 
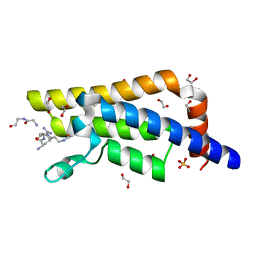 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) complexed with Histone H4-K(ac)5 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, Histone H4, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells.
J Mol Cell Biol, 8, 2016
|
|
2A7Y
 
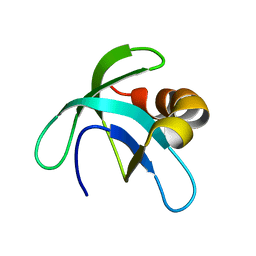 | | Solution Structure of the Conserved Hypothetical Protein Rv2302 from the Bacterium Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv2302/MT2359 | | Authors: | Buchko, G.W, Kim, C.-Y, Terwilliger, T.C, Kennedy, M.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-06 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein Rv2302 from Mycobacterium tuberculosis.
J.Bacteriol., 188, 2006
|
|
2A15
 
 | | X-ray Crystal Structure of RV0760 from Mycobacterium Tuberculosis at 1.68 Angstrom Resolution | | Descriptor: | HYPOTHETICAL PROTEIN Rv0760c, NICOTINAMIDE | | Authors: | Garen, C.R, Cherney, M.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase
Biochim.Biophys.Acta, 1784, 2008
|
|
6ROG
 
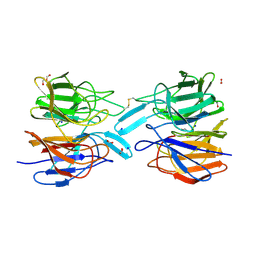 | | Crystal Structure of the KELCH domain of human KEAP1 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, SODIUM ION | | Authors: | Sethi, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bullock, A.N, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of the KELCH domain of human KEAP1
To Be Published
|
|
6SFI
 
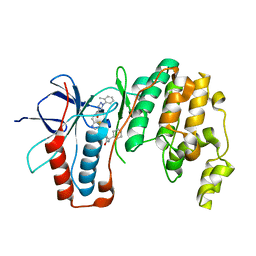 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SFJ
 
 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SE4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor | | Descriptor: | (+)-JD1, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Hassell-Hart, S, Picaud, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Spencer, J, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor
To Be Published
|
|
6SFK
 
 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6T28
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
5KO9
 
 | | Crystal Structure of the SRAP Domain of Human HMCES Protein | | Descriptor: | Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6T6B
 
 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
6T6A
 
 | | Crystal structure of DYRK1A complexed with KuFal319 (compound 11) | | Descriptor: | 4-chloranyl-5~{H}-cyclohepta[b]indol-10-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | [ b ]-Annulated Halogen-Substituted Indoles as Potential DYRK1A Inhibitors.
Molecules, 24, 2019
|
|
8G2H
 
 | | Crystal Structure of PRMT4 with Compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8BB4
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with C3 linker | | Descriptor: | Elongin-B, Elongin-C, WD repeat-containing protein 5, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
8BB5
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with Aryl linker | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
8BIN
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MR21 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-(2-chlorophenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Back-Pocket Optimization of 2-Aminopyrimidine-Based Macrocycles Leads to Potent EPHA2/GAK Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8BIO
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MRAL5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-5-(trifluoromethyl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Lucic, A, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
7Z71
 
 | | Crystal structure of p63 DBD in complex with darpin C14 | | Descriptor: | Darpin C14, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z73
 
 | | Crystal structure of p63 tetramerization domain in complex with darpin 8F1 | | Descriptor: | Darpin 8F1, Isoform 2 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
6SYP
 
 | | Human DHODH bound to inhibitor IPP/CNRS-A017 | | Descriptor: | 2-[4-[2,6-bis(fluoranyl)phenoxy]-5-methyl-3-propan-2-yloxy-pyrazol-1-yl]-5-cyclopropyl-3-fluoranyl-pyridine, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kraemer, A, Janin, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor
Eur.J.Med.Chem., 208, 2020
|
|
5LBA
 
 | | Crystal structure of human RECQL5 helicase in complex with DSPL fragment(1-cyclohexyl-3-(oxolan-2-ylmethyl)urea, SGC - Diamond XChem I04-1 fragment screening. | | Descriptor: | 1-cyclohexyl-3-[[(2~{R})-oxolan-2-yl]methyl]urea, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, ... | | Authors: | Newman, J.A, Aitkenhead, H, Talon, R, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human RECQL5 helicase in complex with 3D fragment (1-cyclohexyl-3-(oxolan-2-ylmethyl)urea)
To be published
|
|
7Z72
 
 | | Crystal structure of p63 SAM in complex with darpin A5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Darpin A5, Isoform 9 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
6RAA
 
 | | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003 | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Arrowsmith, C, Knapp, S, Bountra, C, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003
To Be Published
|
|
4WCI
 
 | | Crystal structure of the 1st SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide (aa 378-393) from human RIN3 | | Descriptor: | CD2-associated protein, Ras and Rab interactor 3, SULFATE ION | | Authors: | Rouka, E, Simister, P.C, Janning, M, Kirsch, K.H, Krojer, T, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
2AQ6
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with pyridoxal 5'-phosphate at 1.7 a resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
