6Z84
 
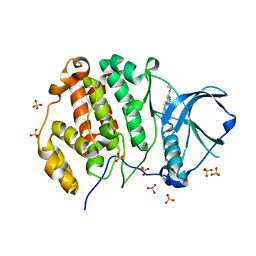 | | CK2 alpha bound to chemical probe SGC-CK2-1 derivative | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[1-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]indol-6-yl]ethanamide | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
6Y57
 
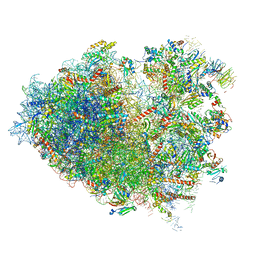 | | Structure of human ribosome in hybrid-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-25 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
3EKZ
 
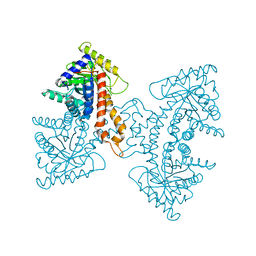 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
3VGA
 
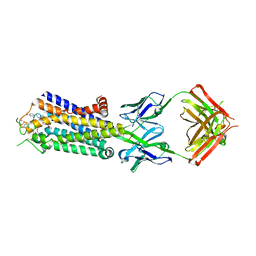 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, antibody fab fragment heavy chain, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
3EJ3
 
 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | Descriptor: | ACETATE ION, Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase, ... | | Authors: | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | Deposit date: | 2008-09-17 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3EJ9
 
 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | Descriptor: | Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase | | Authors: | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | Deposit date: | 2008-09-17 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3EKL
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
3VAP
 
 | | Synthesis and SAR Studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off target kinase selectivity | | Descriptor: | 3-(1-{2-[(3-fluoropyridinium-4-yl)amino]-2-oxoethyl}-1H-pyrazol-4-yl)-6-methyl-8-[(3-{[(1R,3R)-3-methylpiperidinium-1-yl]methyl}-1,2-thiazol-5-yl)amino]imidazo[1,2-a]pyrazin-1-ium, Aurora kinase A | | Authors: | Voss, M.E, Rainka, M.P, Fleming, M, Peterson, L.H, Belanger, D.B, Siddiqui, M.A, Hruza, A, Voigt, J, Basso, A.D, Gray, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Synthesis and SAR studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off-target kinase selectivity.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3UXL
 
 | |
3UXE
 
 | | Design, Synthesis and Biological Evaluation of Potent Quinoline and Pyrroloquinoline Ammosamide Analogues as Inhibitors for Quinone Reductase 2 | | Descriptor: | 8-amino-7-chloro-1-methyl-6-(methylideneamino)-2-oxo-1,2-dihydropyrrolo[4,3,2-de]quinoline-4-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Reddy, N.P, Jensen, K.C, Mesecar, A.D, Fanwick, P.E, Cushman, M. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of potent quinoline and pyrroloquinoline ammosamide analogues as inhibitors of quinone reductase 2.
J.Med.Chem., 55, 2012
|
|
3ZUX
 
 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MERCURY (II) ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
3ZUY
 
 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
3ZCW
 
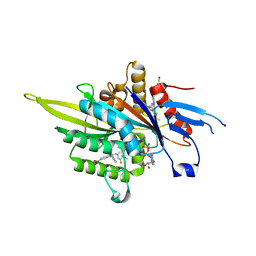 | | Eg5 - New allosteric binding site | | Descriptor: | (2E)-2-(3-fluoranyl-4-methoxy-phenyl)imino-1-[[2-(trifluoromethyl)phenyl]methyl]-3H-benzimidazole-5-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ulaganathan, V, Talapatra, S.K, Kozielski, F, Pannifer, A.D. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Structural Insights Into a Unique Inhibitor Binding Pocket in Kinesin Spindle Protein.
J.Am.Chem.Soc., 135, 2013
|
|
3E9S
 
 | |
3G5E
 
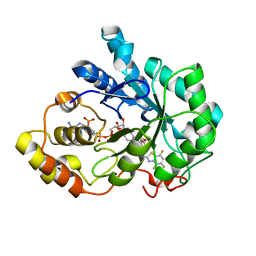 | | Human aldose reductase complexed with IDD 740 inhibitor | | Descriptor: | 2-(3-((4,5,7-trifluorobenzo[d]thiazol-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridin-1-yl)acetic acid, Aldose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Podjarny, A.D, Van Zandt, M.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of [3-(4,5,7-trifluoro-benzothiazol-2-ylmethyl)-pyrrolo[2,3-b]pyridin-1-yl]acetic acids as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3G5M
 
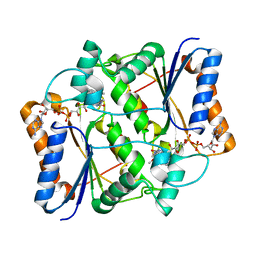 | | Synthesis of Casimiroin and Optimization of Its Quinone Reductase 2 and Aromatase Inhibitory activity | | Descriptor: | 6-methoxy-9-methyl[1,3]dioxolo[4,5-h]quinolin-8(9H)-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Maiti, A, Sturdy, M, Marler, L, Pegan, S.D, Mesecar, A.D, Pezzuto, J.M, Cushman, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Synthesis of casimiroin and optimization of its quinone reductase 2 and aromatase inhibitory activities.
J.Med.Chem., 52, 2009
|
|
3GAM
 
 | | Synthesis of Casimiroin and Optimization of Its Quinone Reductase 2 and Aromatase Inhibitory activity | | Descriptor: | 5,8-dimethoxy-1,4-dimethylquinolin-2(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Sturdy, M, Pegan, S.D, Maiti, A, Marler, L, Mesecar, A.D, Pezzuto, J.M, Cushman, M. | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Synthesis of casimiroin and optimization of its quinone reductase 2 and aromatase inhibitory activities.
J.Med.Chem., 52, 2009
|
|
3ZTT
 
 | | Crystal structure of pneumococcal surface antigen PsaA with manganese | | Descriptor: | MANGANESE (II) ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | McDevitt, C.A, Ogunniyi, A.D, Valkov, E, Lawrence, M.C, Kobe, B, McEwan, A.G, Paton, J.C. | | Deposit date: | 2011-07-12 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular mechanism for bacterial susceptibility to zinc.
PLoS Pathog., 7, 2011
|
|
3UXH
 
 | | Design, Synthesis and Biological Evaluation of Potetent Quinoline and Pyrroloquinoline Ammosamide Analogues as Inhibitors of Quinone Reductase 2 | | Descriptor: | 6,8-diamino-7-chloro-1-methyl-2-oxo-1,2-dihydropyrrolo[4,3,2-de]quinoline-4-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Cushman, M, Mesecar, A.D, Fanwick, P.E, Narasimha, R, Jensen, K.C. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design, synthesis, and biological evaluation of potent quinoline and pyrroloquinoline ammosamide analogues as inhibitors of quinone reductase 2.
J.Med.Chem., 55, 2012
|
|
3DJF
 
 | | Crystal Structure of Schistosoma mansoni Purine Nucleoside Phosphorylase in a complex with BCX-34 | | Descriptor: | 2-amino-7-(pyridin-3-ylmethyl)-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Purine-nucleoside phosphorylase, ... | | Authors: | Postigo, M.P, Pereira, H.M, Oliva, G, Andricopulo, A.D. | | Deposit date: | 2008-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural basis for selective inhibition of purine nucleoside phosphorylase from Schistosoma mansoni: kinetic and structural studies.
Bioorg.Med.Chem., 18, 2010
|
|
3X3B
 
 | | Crystal structure of the light-driven sodium pump KR2 in acidic state | | Descriptor: | DI(HYDROXYETHYL)ETHER, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
3X3C
 
 | | Crystal structure of the light-driven sodium pump KR2 in neutral state | | Descriptor: | OLEIC ACID, RETINAL, Sodium pumping rhodopsin | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
4ARD
 
 | | Structure of the immature retroviral capsid at 8A resolution by cryo- electron microscopy | | Descriptor: | CAPSID PROTEIN P27 | | Authors: | Bharat, T.A.M, Davey, N.E, Ulbrich, P, Riches, J.D, Marco, A.D, Rumlova, M, Sachse, C, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the Immature Retroviral Capsid at 8A Resolution by Cryo-Electron Microscopy.
Nature, 487, 2012
|
|
4APS
 
 | | Crystal structure of a POT family peptide transporter in an inward open conformation. | | Descriptor: | CADMIUM ION, DI-OR TRIPEPTIDE H+ SYMPORTER | | Authors: | Solcan, N, Kwok, J, Fowler, P.W, Cameron, A.D, Drew, D, Iwata, S, Newstead, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternating Access Mechanism in the Pot Family of Oligopeptide Transporters.
Embo J., 31, 2012
|
|
