8GVJ
 
 | | Crystal structure of cMET kinase domain bound by D6808 | | Descriptor: | (1^4Z,5^2E)-6^3-(trifluoromethyl)-5^1,5^6-dihydro-1^1H-8-aza-2(3,6)-quinolina-5(1,3)-pyridazina-1(4,1)-pyrazola-6(1,4)-benzenacyclododecaphane-5^6,7-dione, Hepatocyte growth factor receptor | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2022-09-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
8GZF
 
 | | Crystal Structure of METTL9-SAH | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
8GZE
 
 | | Crystal Structure of human METTL9-SAH-SLC39A7 peptide complex | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, Zinc transporter SLC39A7 | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
8GTW
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound JZD-26 | | Descriptor: | (2S)-4-(3,4-dichlorophenyl)-1-[(2-oxidanylidene-1H-quinolin-4-yl)carbonyl]-N-[3,3,3-tris(fluoranyl)propyl]piperazine-2-carboxamide, 3C-like proteinase | | Authors: | Su, H.X, Nie, T.Q, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of novel inhibitors against SARS-CoV-2 3CLpro
To Be Published
|
|
8GTV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound JZD-07 | | Descriptor: | 3C-like proteinase, 4-[(2~{S})-4-(3,4-dichlorophenyl)-2-(morpholin-4-ylmethyl)piperazin-1-yl]carbonyl-1~{H}-quinolin-2-one | | Authors: | Su, H.X, Nie, T.Q, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel inhibitors against SARS-CoV-2 3CLpro
To Be Published
|
|
3L0I
 
 | | Complex structure of SidM/DrrA with the wild type Rab1 | | Descriptor: | CHLORIDE ION, DrrA, Ras-related protein Rab-1A, ... | | Authors: | Zhu, Y, Shao, F. | | Deposit date: | 2009-12-10 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural mechanism of host Rab1 activation by the bifunctional Legionella type IV effector SidM/DrrA
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L0M
 
 | |
6I3M
 
 | | eIF2B:eIF2 complex, phosphorylated on eIF2 alpha serine 52. | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Adomavicius, T, Roseman, A.M, Pavitt, G.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-05-22 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The structural basis of translational control by eIF2 phosphorylation.
Nat Commun, 10, 2019
|
|
6I7T
 
 | | eIF2B:eIF2 complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Adomavicius, T, Guaita, M, Roseman, A.M, Pavitt, G.D. | | Deposit date: | 2018-11-17 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.61 Å) | | Cite: | The structural basis of translational control by eIF2 phosphorylation.
Nat Commun, 10, 2019
|
|
6IEG
 
 | | Crystal structure of human MTR4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
3DPC
 
 | |
8ESB
 
 | |
8ESA
 
 | |
8ES9
 
 | | CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM.
Nat Commun, 14, 2023
|
|
8ES7
 
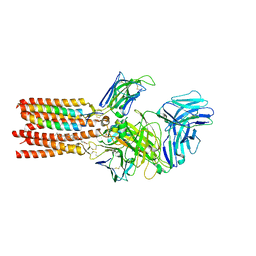 | | CryoEM structure of PN45545 TCR-CD3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM.
Nat Commun, 14, 2023
|
|
8ES8
 
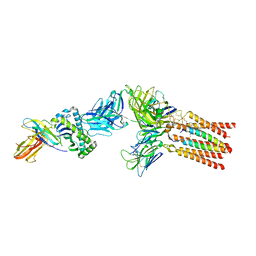 | |
2JXW
 
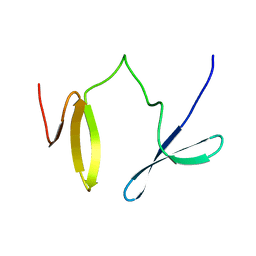 | |
5K6J
 
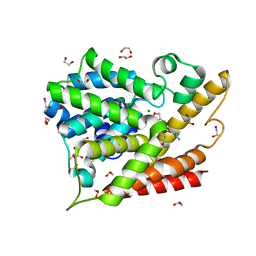 | | Human Phospodiesterase 4B in complex with pyridyloxy-benzoxaborole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-[[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl]oxy]-5-chloranyl-2-(4-oxidanylidenepentoxy)pyridine-3-carbonitrile, MAGNESIUM ION, ... | | Authors: | Rock, F.L, Zhou, Y, Sullivan, D. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | To be published
To Be Published
|
|
5YIK
 
 | |
5YLC
 
 | | Crystal Structure of MCR-1 Catalytic Domain | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
5YIJ
 
 | | Structure of a Legionella effector with substrates | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | Authors: | Feng, Y, Mu, Y, Wang, H. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
7FFM
 
 | | Human serum transferrin with five osmium binding sites | | Descriptor: | MALONATE ION, NITRILOTRIACETIC ACID, OSMIUM ION, ... | | Authors: | Wang, M, Sun, H. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
5DOI
 
 | | Crystal structure of Tetrahymena p45N and p19 | | Descriptor: | Telomerase associated protein p45, Telomerase-associated protein 19 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
5YLE
 
 | | MCR-1 complex with ethanolamine (ETA) | | Descriptor: | ETHANOLAMINE, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
