6DS6
 
 | |
6E86
 
 | |
8HUX
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
7WMW
 
 | |
7DUY
 
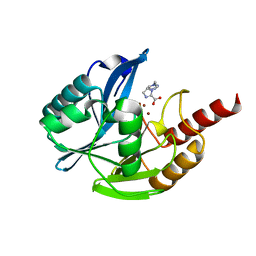 | | Crystal structure of VIM-2 MBL in complex with 1-(2-(1H-1,2,3-triazol-1-yl)ethyl)-1H-imidazole-2-carboxylic acid | | Descriptor: | 1-[2-(1,2,3-triazol-1-yl)ethyl]imidazole-2-carboxylic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure-guided optimization of 1H-imidazole-2-carboxylic acid derivatives affording potent VIM-Type metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 228, 2022
|
|
7DV0
 
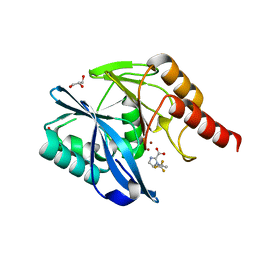 | |
7DUX
 
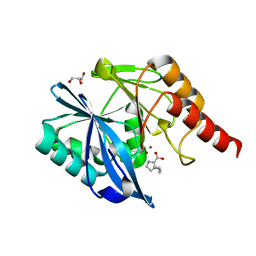 | |
7WMY
 
 | |
7WMX
 
 | |
7DV1
 
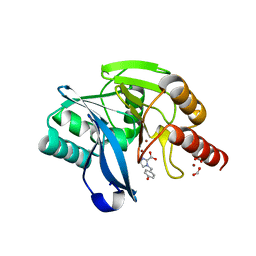 | |
7DUZ
 
 | |
7DU1
 
 | |
7DUE
 
 | |
7DUB
 
 | |
7WMZ
 
 | |
7DYY
 
 | |
7DYZ
 
 | |
7DZ1
 
 | |
7DZ0
 
 | |
9J9J
 
 | | Nitrophenol monooxygenase RsPNPA from Rhodococcus sp. 21391 | | Descriptor: | 4-nitrophenol 4-monooxygenase/4-nitrocatechol 2-monooxygenase, oxygenase component | | Authors: | Yang, J, Li, R, Lin, S, Long, L. | | Deposit date: | 2024-08-22 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biodegradation of p-nitrophenol by Rhodococcus sp. 21391 unveils a two-component p-nitrophenol monooxygenase with broad substrate specificity.
Microb Cell Fact, 24, 2025
|
|
7X83
 
 | | Cryo-EM structure of the TMEM106B fibril from normal elder | | Descriptor: | Transmembrane protein 106B | | Authors: | Xia, W.C, Zhao, Q.Y, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
7X84
 
 | | Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia | | Descriptor: | Transmembrane protein 106B | | Authors: | Zhao, Q.Y, Xia, W.C, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
3HIA
 
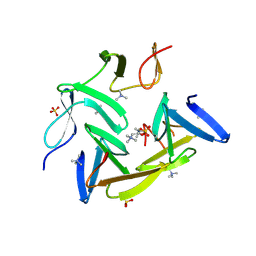 | | Crystal structure of the choline binding domain of Spr1274 in Streptococcus pneumoniae | | Descriptor: | Choline binding protein, PHOSPHOCHOLINE, SULFATE ION | | Authors: | Zhang, Z.-Y, Li, W.-Z, Jiang, Y.-L, Bao, R, Zhou, C.-Z, Chen, Y.-X. | | Deposit date: | 2009-05-19 | | Release date: | 2009-08-04 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of the choline-binding domain of Spr1274 in Streptococcus pneumoniae.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7PO5
 
 | |
7PNM
 
 | |
