8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
4QQC
 
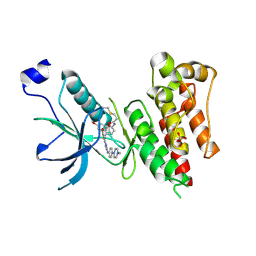 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R5S
 
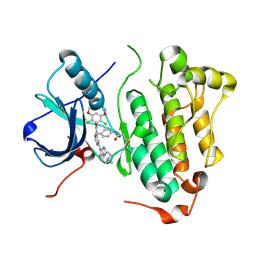 | | Crystal structure of EGFR 696-1022 L858R in complex with FIIN-3 | | Descriptor: | Epidermal growth factor receptor, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R6V
 
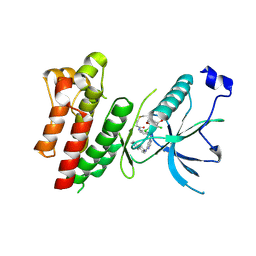 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7FAH
 
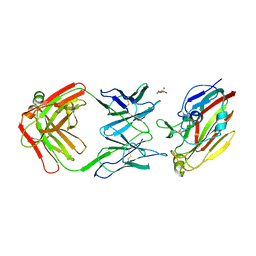 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
6V4O
 
 | |
6V4N
 
 | |
6LDQ
 
 | |
6NV1
 
 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, ... | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-02-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
7JVN
 
 | |
7JVM
 
 | |
4D1E
 
 | | THE CRYSTAL STRUCTURE OF HUMAN MUSCLE ALPHA-ACTININ-2 | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Pinotsis, N, Salmazo, A, Sjoeblom, B, Gkougkoulia, E, Djinovic-Carugo, K. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure and Regulation of Human Muscle Alpha-Actinin
Cell(Cambridge,Mass.), 159, 2014
|
|
6L8T
 
 | |
6MD9
 
 | |
6MDA
 
 | |
5EPO
 
 | | The three-dimensional structure of Clostridium absonum 7alpha-hydroxysteroid dehydrogenase | | Descriptor: | 7-alpha-hydroxysteroid deydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lou, D, Wang, B, Wang, F. | | Deposit date: | 2015-11-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Clostridium absonum 7 alpha-hydroxysteroid dehydrogenase: new insights into the conserved arginines for NADP(H) recognition
Sci Rep, 6, 2016
|
|
8KH8
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH6
 
 | | Crystal structure of FGFR4 kinase domain with 8r | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[5-bromanyl-6-(hydroxymethyl)-3-methoxy-pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH9
 
 | | Crystal structure of FGFR4(V550M) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH7
 
 | | Crystal structure of FGFR4 kinase domain with 8zc | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
7KHF
 
 | |
7KFK
 
 | |
7WM0
 
 | |
7WLY
 
 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
