7C29
 
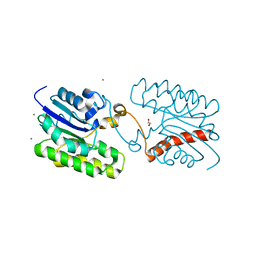 | | Esterase CrmE10 mutant-D178A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
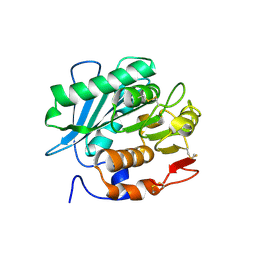
7YME
 
 | |
6YN9
 
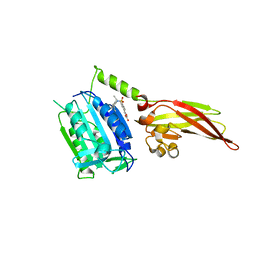 | | MALT1(329-728) in complex with a sulfonamide containing compound | | Descriptor: | 5-[4-[(2,6-diethylphenyl)sulfamoyl]-3-methyl-phenyl]furan-3-carboxylic acid, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
6G3G
 
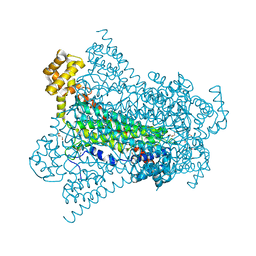 | | Crystal structure of EDDS lyase in complex with succinate | | Descriptor: | Argininosuccinate lyase, DI(HYDROXYETHYL)ETHER, SUCCINIC ACID | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
3TL5
 
 | | Discovery of GDC-0980: a Potent, Selective, and Orally Available Class I Phosphatidylinositol 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Kinase Inhibitor for the Treatment of Cancer | | Descriptor: | (2S)-1-(4-{[2-(2-aminopyrimidin-5-yl)-7-methyl-4-(morpholin-4-yl)thieno[3,2-d]pyrimidin-6-yl]methyl}piperazin-1-yl)-2-hydroxypropan-1-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2011-08-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available Class I Phosphatidylinositol 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Kinase Inhibitor (GDC-0980) for the Treatment of Cancer.
J.Med.Chem., 54, 2011
|
|
6YPZ
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
5CDM
 
 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
6YQ6
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6G3E
 
 | | Crystal structure of EDDS lyase in complex with formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, SODIUM ION | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
7F2L
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 18a | | Descriptor: | (~{E})-4-[9-[(4-fluorophenyl)methoxy]-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10111427 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
5TZR
 
 | | GPR40 in complex with partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, Free fatty acid receptor 1,Endolysin,Free fatty acid receptor 1, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7F2M
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 18d | | Descriptor: | (~{Z})-4-[9-[(4-fluorophenyl)methoxy]-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20004153 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
7F2K
 
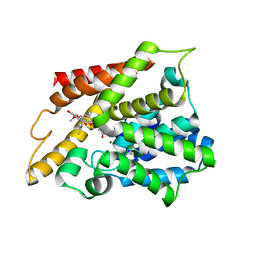 | | Crystal structure of PDE4D catalytic domain complexed with compound 17a | | Descriptor: | (~{E})-4-[8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-9-oxidanyl-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10001969 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
6YQ3
 
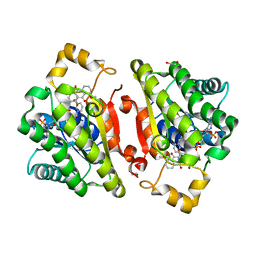 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
4XS2
 
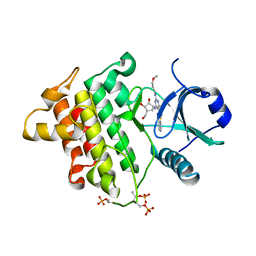 | | Irak4-inhibitor co-structure | | Descriptor: | (1R,2S,3R,5R)-3-({5-(1,3-benzothiazol-2-yl)-6-chloro-2-[(3-methoxypropyl)amino]pyrimidin-4-yl}amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery and hit-to-lead optimization of 2,6-diaminopyrimidine inhibitors of interleukin-1 receptor-associated kinase 4.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3KJ4
 
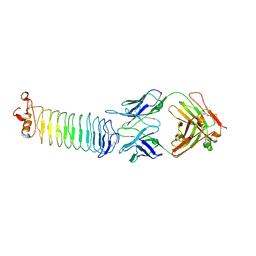 | | Structure of rat Nogo receptor bound to 1D9 antagonist antibody | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment 1D9 heavy chain, ... | | Authors: | Silvian, L.F. | | Deposit date: | 2009-11-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Resolution of disulfide heterogeneity in Nogo receptor 1 fusion proteins by molecular engineering.
Biotechnol Appl Biochem, 57, 2010
|
|
8ILQ
 
 | | Structure of SFTSV Gn-Gc heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
8I4T
 
 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
8TBP
 
 | | HLA-DRB1*15:01 in complex with smith antigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ting, Y.T, Broury, A, Ooi, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.12621117 Å) | | Cite: | Smith-specific regulatory T cells halt the progression of lupus nephritis.
Nat Commun, 15, 2024
|
|
2L5G
 
 | | Co-ordinates and 1H, 13C and 15N chemical shift assignments for the complex of GPS2 53-90 and SMRT 167-207 | | Descriptor: | G protein pathway suppressor 2, Putative uncharacterized protein NCOR2 | | Authors: | Oberoi, J, Yang, J, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7FBB
 
 | | De novo design protein D12 with MBP tag | | Descriptor: | Maltodextrin-binding protein,de novo designed protein D12 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBD
 
 | | De novo design protein D53 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D53 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6YN8
 
 | | Human MALT1(334-719) in complex with a tetrazole containing compound | | Descriptor: | 3-azanyl-3-methyl-~{N}-[(3~{R})-4-oxidanylidene-5-[[4-[2-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]-2,3-dihydro-1,5-benzoxazepin-3-yl]butanamide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
7FBC
 
 | | De novo design protein D22 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D22 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6G3D
 
 | | Crystal structure of Native EDDS lyase | | Descriptor: | Argininosuccinate lyase | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
