2L3H
 
 | | NMR Structure in a Membrane Environment Reveals Putative Amyloidogenic Regions of the SEVI Precursor Peptide PAP248-286 | | Descriptor: | Prostatic acid phosphatase | | Authors: | Ramamoorthy, A, Nanga, R, Brender, J, Vivekanandan, S, Popovych, N. | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure in a membrane environment reveals putative amyloidogenic regions of the SEVI precursor peptide PAP(248-286).
J.Am.Chem.Soc., 131, 2009
|
|
4HPY
 
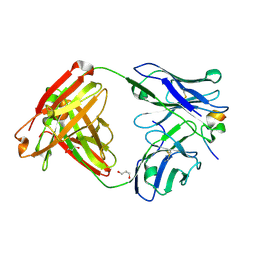 | | Crystal structure of RV144-elicited antibody CH59 in complex with V2 peptide | | Descriptor: | CH59 Fab heavy chain, CH59 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
4I3R
 
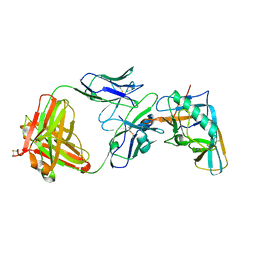 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
3WLL
 
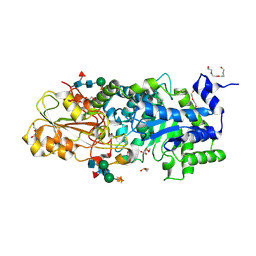 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with PEG400 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
2LDM
 
 | |
3TIH
 
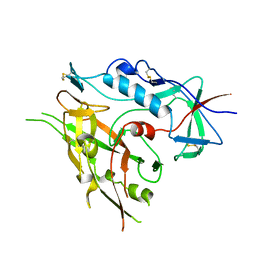 | |
4HHG
 
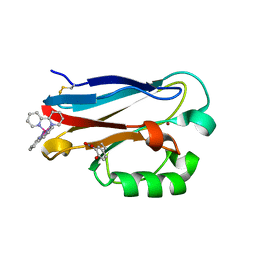 | | Crystal structure of the Pseudomonas aeruginosa azurin, RuH107NO YOH109 | | Descriptor: | Azurin, COPPER (II) ION, DELTA-BIS(2,2'-BIPYRIDINE)IMIDAZOLE RUTHENIUM (II) | | Authors: | Herrera, N, Warren, J.J, Gray, H.B. | | Deposit date: | 2012-10-09 | | Release date: | 2012-11-21 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Electron Flow through Nitrotyrosinate in Pseudomonas aeruginosa Azurin.
J.Am.Chem.Soc., 135, 2013
|
|
3WLN
 
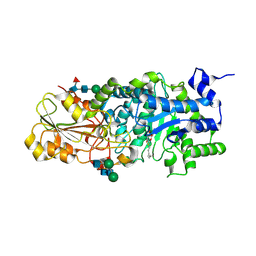 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with octyl-S-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLK
 
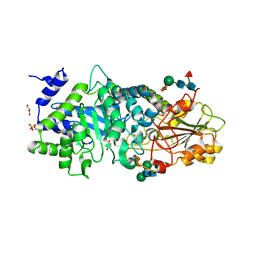 | |
3WLM
 
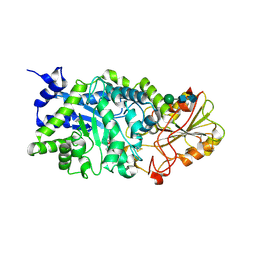 | |
4H8S
 
 | | Crystal structure of human APPL2BARPH domain | | Descriptor: | DCC-interacting protein 13-beta | | Authors: | Martin, J.L, King, G.J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane Curvature Protein Exhibits Interdomain Flexibility and Binds a Small GTPase.
J.Biol.Chem., 287, 2012
|
|
4HGE
 
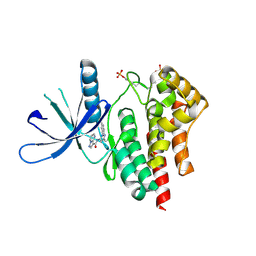 | | JAK2 kinase (JH1 domain) in complex with compound 8 | | Descriptor: | N-[1-(3-chlorophenyl)-3-methyl-1H-pyrazol-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent and selective pyrazolopyrimidine janus kinase 2 inhibitors.
J.Med.Chem., 55, 2012
|
|
3U1S
 
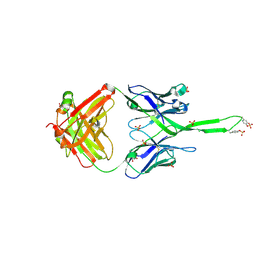 | | Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Fab PGT145 Heavy chain, Fab PGT145 Light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Diwanji, D, Burton, D.R, Wilson, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3TKY
 
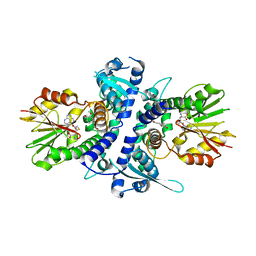 | | Monolignol o-methyltransferase (momt) | | Descriptor: | (Iso)eugenol O-methyltransferase, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bhuiya, M.W, Liu, C.J. | | Deposit date: | 2011-08-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | An engineered monolignol 4-o-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis.
Plant Cell, 24, 2012
|
|
4I5C
 
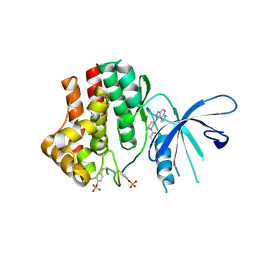 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-3-[(3R)-3-(pyrrolo[2,3-b][1,2,3]triazolo[4,5-d]pyridin-1(6H)-yl)piperidin-1-yl]propanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazolo-pyrrolopyridines as inhibitors of Janus kinase 1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2KRE
 
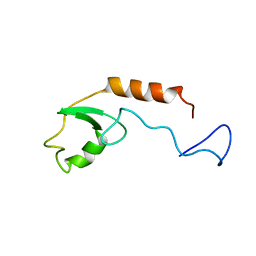 | |
4HHW
 
 | |
3WLO
 
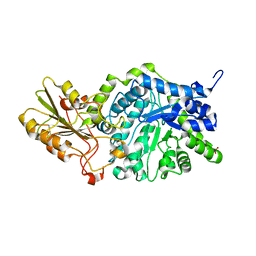 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, SULFATE ION, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLP
 
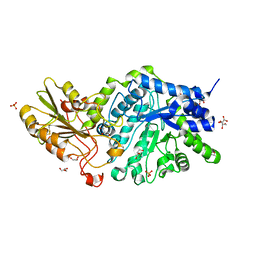 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 1-thio-beta-D-glucopyranose-(1-6)-methyl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLI
 
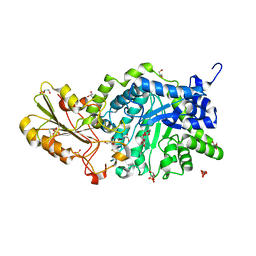 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
4HPO
 
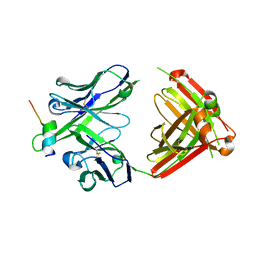 | | Crystal structure of RV144-elicited antibody CH58 in complex with V2 peptide | | Descriptor: | CH58 Fab heavy chain, CH58 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
3U46
 
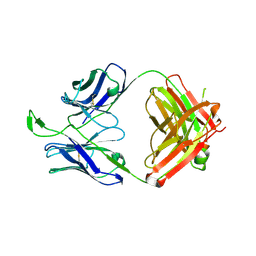 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
4I3V
 
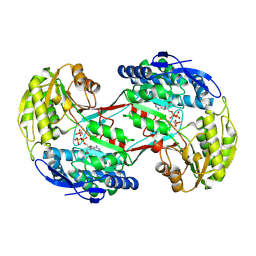 | |
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
4I3U
 
 | |
